Evolutionary and functional analysis of mulberry type III polyketide synthases
- PMID: 27487946
- PMCID: PMC4973071
- DOI: 10.1186/s12864-016-2843-7
Evolutionary and functional analysis of mulberry type III polyketide synthases
Abstract
Background: Type III polyketide synthases are important for the biosynthesis of flavonoids and various plant polyphenols. Mulberry plants have abundant polyphenols, but very little is known about the mulberry type III polyketide synthase genes. An analysis of these genes may provide new targets for genetic improvement to increase relevant secondary metabolites and enhance the plant tolerance to biotic and abiotic stresses.
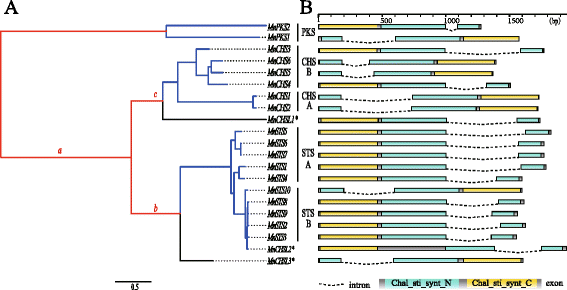
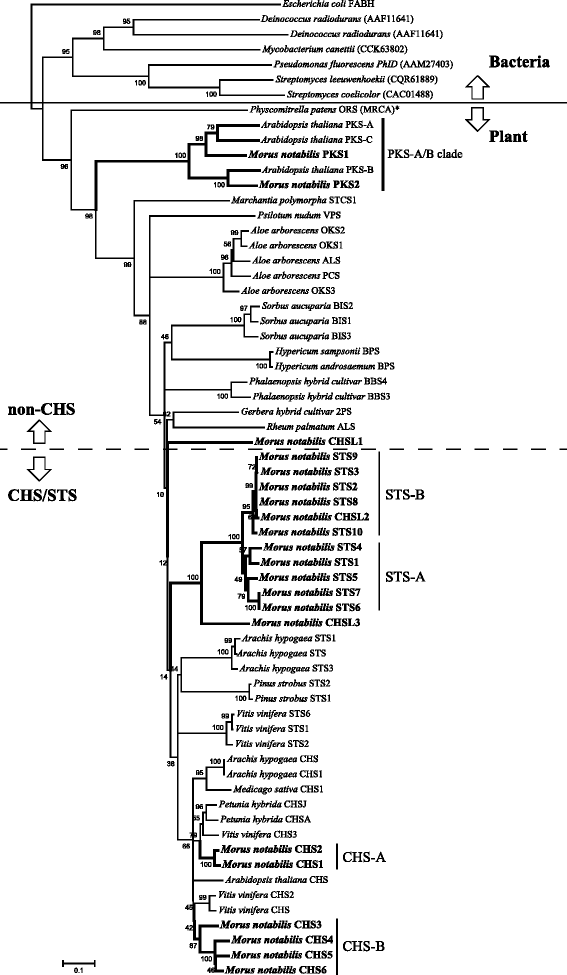
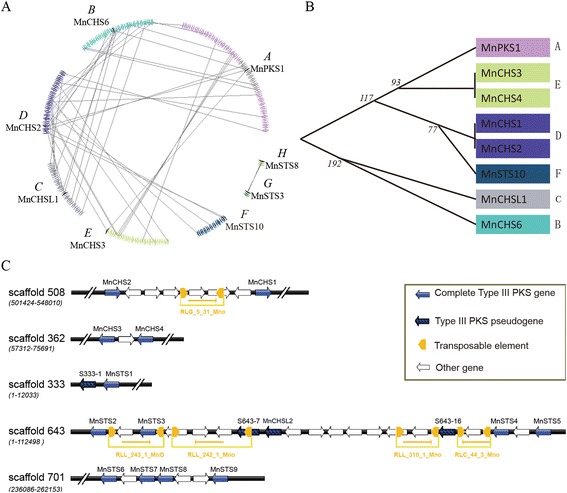
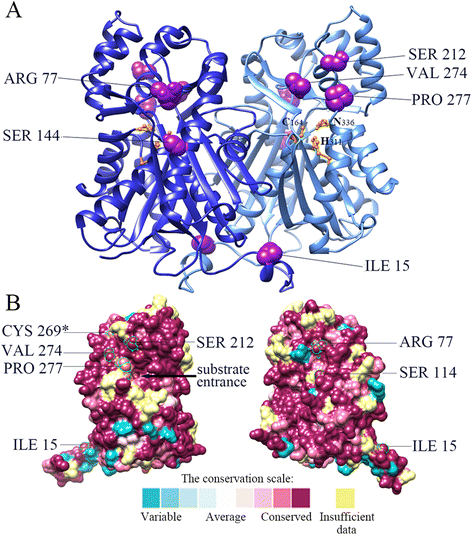
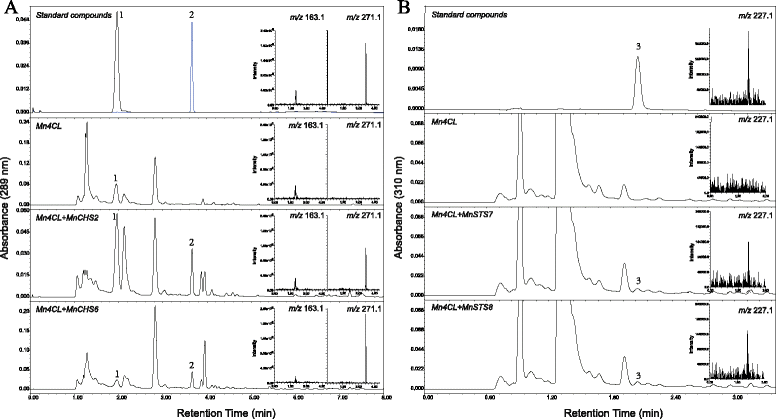
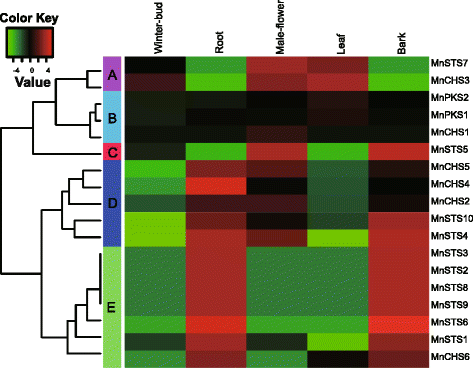
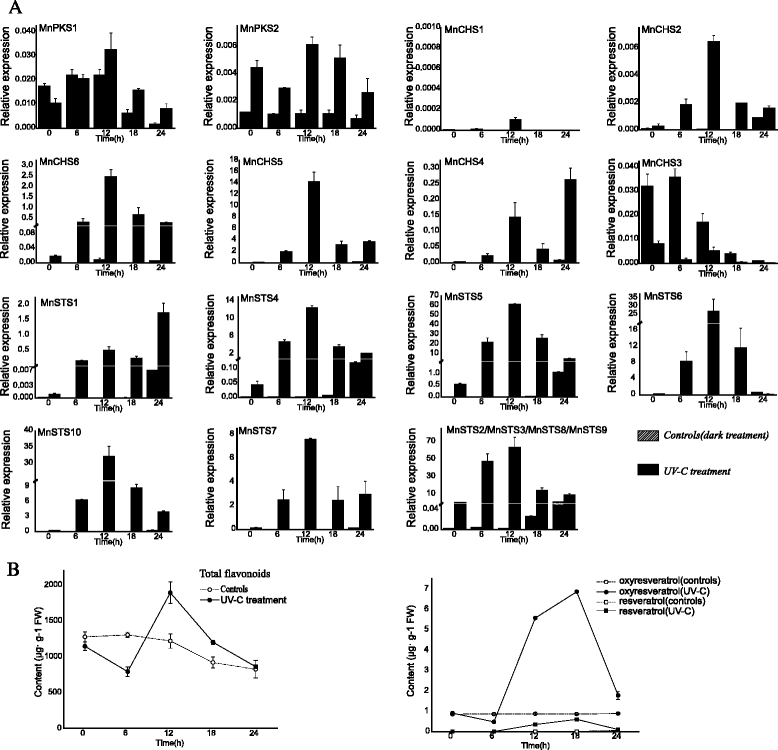
Results: Eighteen genes encoding type III polyketide synthases were identified, including six chalcone synthases (CHS), ten stilbene synthases (STS), and two polyketide synthases (PKS). Functional characterization of four genes representing most of the MnCHS and MnSTS genes by coexpression with 4-Coumaroyl-CoA ligase in Escherichia coli indicated that their products were able to catalyze p-coumaroyl-CoA and malonyl-CoA to generate naringenin and resveratrol, respectively. Microsynteny analysis within mulberry indicated that segmental and tandem duplication events contributed to the expansion of the MnCHS family, while tandem duplications were mainly responsible for the generation of the MnSTS genes. Combining the evolution and expression analysis results of the mulberry type III PKS genes indicated that MnCHS and MnSTS genes evolved mainly under purifying selection to maintain their original functions, but transcriptional subfunctionalization occurred during long-term species evolution. Moreover, mulberry leaves can rapidly accumulated oxyresveratrol after UV-C irradiation, suggesting that resveratrol was converted to oxyresveratrol.
Conclusions: Characterizing the functions and evolution of mulberry type III PKS genes is crucial for advancing our understanding of these genes and providing the basis for further studies on the biosynthesis of relevant secondary metabolites in mulberry plants.
Keywords: Chalcone synthase; Evolutionary analysis; Functional analysis; Gene expression; Mulberry; Stilbene synthase.
Figures
References
-
- Schnee S, Viret O, Gindro K. Role of stilbenes in the resistance of grapevine to powdery mildew. Physiol Mol Plant Pathol. 2008;72:128–133. doi: 10.1016/j.pmpp.2008.07.002. - DOI
-
- Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol J. 2007;2(10):1214–1234. doi: 10.1002/biot.200700084. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

