Norovirus Whole-Genome Sequencing by SureSelect Target Enrichment: a Robust and Sensitive Method
- PMID: 27487952
- PMCID: PMC5035417
- DOI: 10.1128/JCM.01052-16
Norovirus Whole-Genome Sequencing by SureSelect Target Enrichment: a Robust and Sensitive Method
Abstract
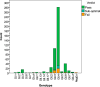
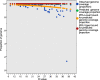
Norovirus full-genome sequencing is challenging due to sequence heterogeneity among genomes. Previous methods have relied on PCR amplification, which is problematic due to primer design, and transcriptome sequencing (RNA-Seq), which nonspecifically sequences all RNA, including host and bacterial RNA, in stool specimens. Target enrichment uses a panel of custom-designed 120-mer RNA baits that are complementary to all publicly available norovirus sequences, with multiple baits targeting each position of the genome, which overcomes the challenge of primer design. Norovirus genomes are enriched from stool RNA extracts to minimize the sequencing of nontarget RNA. SureSelect target enrichment and Illumina sequencing were used to sequence full genomes from 507 norovirus-positive stool samples with reverse transcription-real-time PCR cycle threshold (CT) values of 10 to 43. Sequencing on an Illumina MiSeq system in batches of 48 generated, on average, 81% on-target reads per sample and 100% genome coverage with >12,000-fold read depth. Samples included genotypes GI.1, GI.2, GI.3, GI.6, GI.7, GII.1, GII.2, GII.3, GII.4, GII.5, GII.6, GII.7, GII.13, GII.14, and GII.17. When outliers were accounted for, we generated >80% genome coverage for all positive samples, regardless of CT values. A total of 164 samples were tested in parallel with conventional PCR genotyping of the capsid shell domain; 164/164 samples were successfully sequenced, compared to 158/164 samples that were amplified by PCR. Four of the samples that failed capsid PCR analysis had low titers, which suggests that target enrichment is more sensitive than gel-based PCR. Two samples failed PCR due to primer mismatches; target enrichment uses multiple baits targeting each position, thus accommodating sequence heterogeneity among norovirus genomes.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

