Adding energy minimization strategy to peptide-design algorithm enables better search for RNA-binding peptides: Redesigned λ N peptide binds boxB RNA
- PMID: 27487990
- PMCID: PMC5314887
- DOI: 10.1002/jcc.24466
Adding energy minimization strategy to peptide-design algorithm enables better search for RNA-binding peptides: Redesigned λ N peptide binds boxB RNA
Abstract
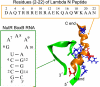
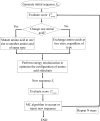
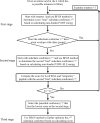
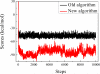
Our previously developed peptide-design algorithm was improved by adding an energy minimization strategy which allows the amino acid sidechains to move in a broad configuration space during sequence evolution. In this work, the new algorithm was used to generate a library of 21-mer peptides which could substitute for λ N peptide in binding to boxB RNA. Six potential peptides were obtained from the algorithm, all of which exhibited good binding capability with boxB RNA. Atomistic molecular dynamics simulations were then conducted to examine the ability of the λ N peptide and three best evolved peptides, viz. Pept01, Pept26, and Pept28, to bind to boxB RNA. Simulation results demonstrated that our evolved peptides are better at binding to boxB RNA than the λ N peptide. Sequence searches using the old (without energy minimization strategy) and new (with energy minimization strategy) algorithms confirm that the new algorithm is more effective at finding good RNA-binding peptides than the old algorithm. © 2016 Wiley Periodicals, Inc.
Keywords: RNA tethering; RNA-binding peptide; energy minimization strategy; peptide-design algorithm; λ N peptide-boxB RNA.
© 2016 Wiley Periodicals, Inc.
Figures




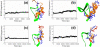
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

