The circadian clock regulates inflammatory arthritis
- PMID: 27488122
- PMCID: PMC5067252
- DOI: 10.1096/fj.201600353R
The circadian clock regulates inflammatory arthritis
Abstract
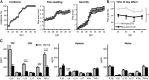
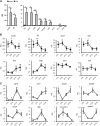
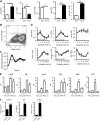
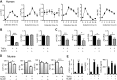
There is strong diurnal variation in the symptoms and severity of chronic inflammatory diseases, such as rheumatoid arthritis. In addition, disruption of the circadian clock is an aggravating factor associated with a range of human inflammatory diseases. To investigate mechanistic links between the biological clock and pathways underlying inflammatory arthritis, mice were administered collagen (or saline as a control) to induce arthritis. The treatment provoked an inflammatory response within the limbs, which showed robust daily variation in paw swelling and inflammatory cytokine expression. Inflammatory markers were significantly repressed during the dark phase. Further work demonstrated an active molecular clock within the inflamed limbs and highlighted the resident inflammatory cells, fibroblast-like synoviocytes (FLSs), as a potential source of the rhythmic inflammatory signal. Exposure of mice to constant light disrupted the clock in peripheral tissues, causing loss of the nighttime repression of local inflammation. Finally, the results show that the core clock proteins cryptochrome (CRY) 1 and 2 repressed inflammation within the FLSs, and provide novel evidence that a CRY activator has anti-inflammatory properties in human cells. We conclude that under chronic inflammatory conditions, the clock actively represses inflammatory pathways during the dark phase. This interaction has exciting potential as a therapeutic avenue for treatment of inflammatory disease.-Hand, L. E., Hopwood, T. W., Dickson, S. H., Walker, A. L., Loudon, A. S. I., Ray, D. W., Bechtold, D. A., Gibbs, J. E. The circadian clock regulates inflammatory arthritis.
Keywords: cryptochrome; diurnal; fibroblast-like synoviocyte; rheumatoid arthritis.
© The Author(s).
Figures


References
-
- Hastings M. H., Maywood E. S., Reddy A. B. (2008) Two decades of circadian time. J. Neuroendocrinol. 20, 812–819 - PubMed
-
- Labrecque N., Cermakian N. (2015) Circadian clocks in the immune system. J. Biol. Rhythms 30, 277–290 - PubMed
-
- Gibbs J. E., Blaikley J., Beesley S., Matthews L., Simpson K. D., Boyce S. H., Farrow S. N., Else K. J., Singh D., Ray D. W., Loudon A. S. (2012) The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 109, 582–587 - PMC - PubMed
-
- Arjona A., Sarkar D. K. (2005) Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J. Immunol. 174, 7618–7624 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

