Predominant expression of Alzheimer's disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts
- PMID: 27488240
- PMCID: PMC4973113
- DOI: 10.1186/s13024-016-0124-1
Predominant expression of Alzheimer's disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts
Erratum in
-
Correction: Predominant expression of Alzheimer's disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts.Mol Neurodegener. 2023 Oct 2;18(1):72. doi: 10.1186/s13024-023-00662-z. Mol Neurodegener. 2023. PMID: 37784204 Free PMC article. No abstract available.
Abstract
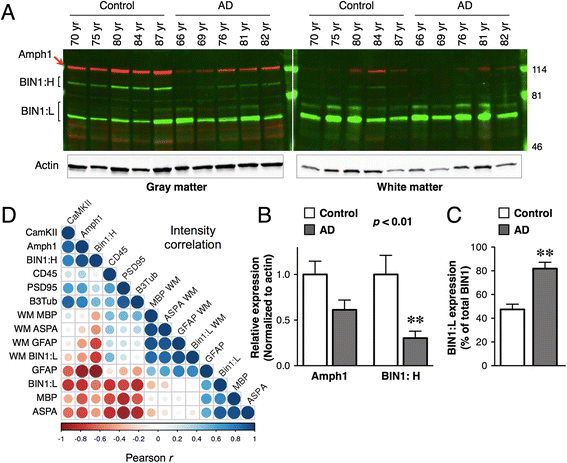
Background: Genome-wide association studies have identified BIN1 within the second most significant susceptibility locus in late-onset Alzheimer's disease (AD). BIN1 undergoes complex alternative splicing to generate multiple isoforms with diverse functions in multiple cellular processes including endocytosis and membrane remodeling. An increase in BIN1 expression in AD and an interaction between BIN1 and Tau have been reported. However, disparate descriptions of BIN1 expression and localization in the brain previously reported in the literature and the lack of clarity on brain BIN1 isoforms present formidable challenges to our understanding of how genetic variants in BIN1 increase the risk for AD.
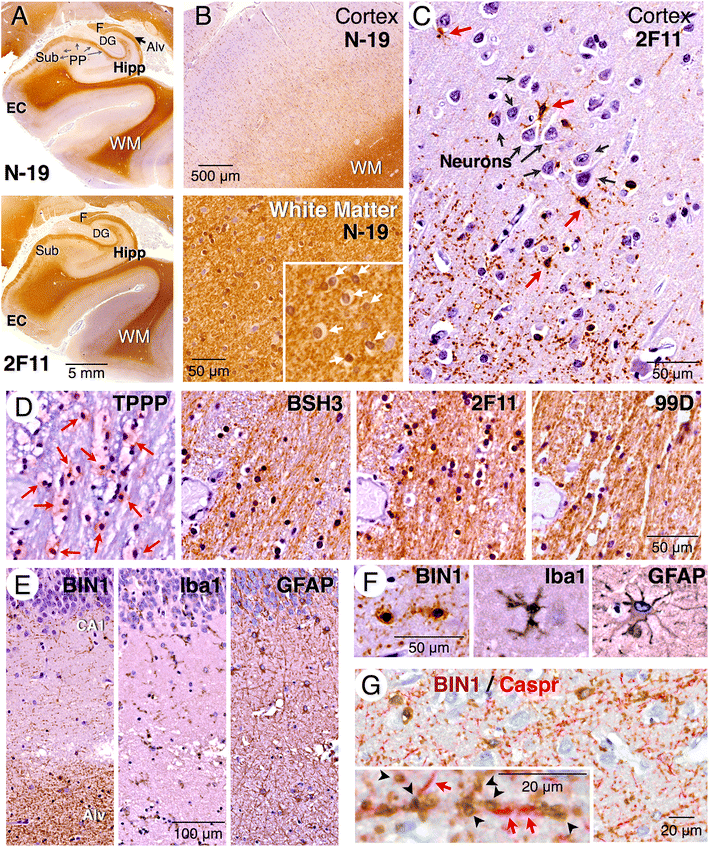
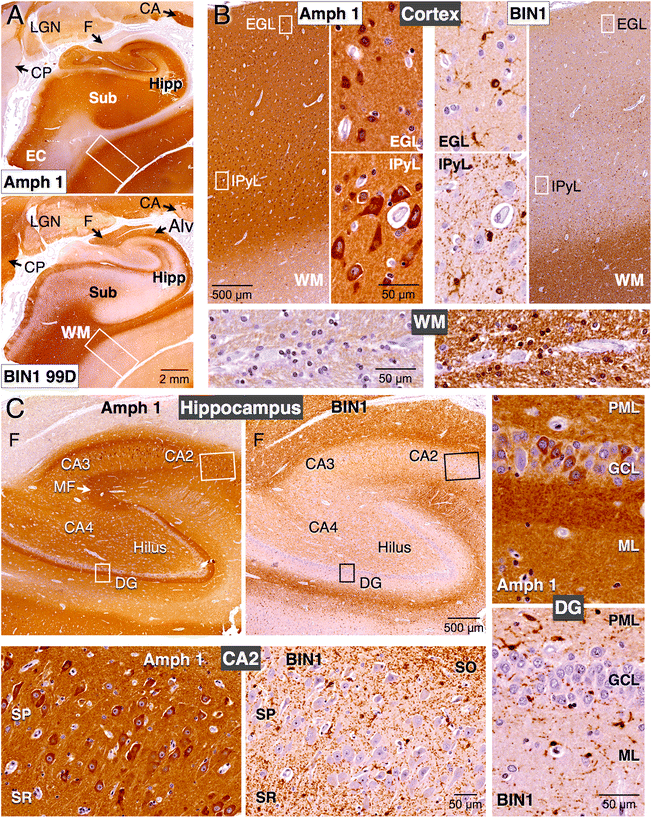
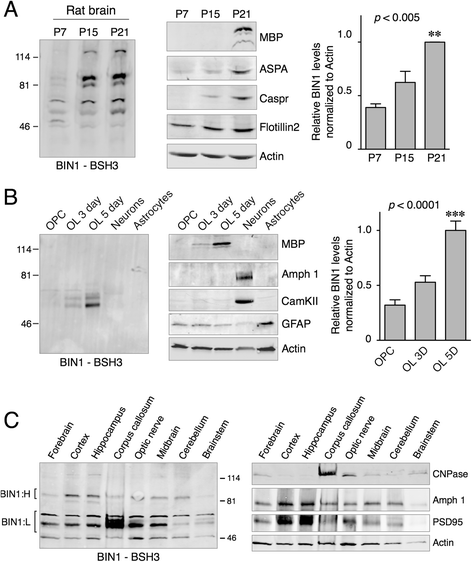
Methods: In this study, we analyzed BIN1 mRNA and protein levels in human brain samples from individuals with or without AD. In addition, we characterized the BIN1 expression and isoform diversity in human and rodent tissue by immunohistochemistry and immunoblotting using a panel of BIN1 antibodies.
Results: Here, we report on BIN1 isoform diversity in the human brain and document alterations in the levels of select BIN1 isoforms in individuals with AD. In addition, we report striking BIN1 localization to white matter tracts in rodent and the human brain, and document that the large majority of BIN1 is expressed in mature oligodendrocytes whereas neuronal BIN1 represents a minor fraction. This predominant non-neuronal BIN1 localization contrasts with the strict neuronal expression and presynaptic localization of the BIN1 paralog, Amphiphysin 1. We also observe upregulation of BIN1 at the onset of postnatal myelination in the brain and during differentiation of cultured oligodendrocytes. Finally, we document that the loss of BIN1 significantly correlates with the extent of demyelination in multiple sclerosis lesions.
Conclusion: Our study provides new insights into the brain distribution and cellular expression of an important risk factor associated with late-onset AD. We propose that efforts to define how genetic variants in BIN1 elevate the risk for AD would behoove to consider BIN1 function in the context of its main expression in mature oligodendrocytes and the potential for a role of BIN1 in the membrane remodeling that accompanies the process of myelination.
Keywords: Alternative splicing; Alzheimer’s disease; Amphiphysin 1; BIN1; Immunohistochemistry; Isoform diversity; Late-onset Alzheimer’s disease; Multiple sclerosis; Myelination; Oligodendrocyte.
Figures





References
-
- Butler MH, David C, Ochoa GC, Freyberg Z, Daniell L, Grabs D, Cremona O, De Camilli P. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

