Changes in Structure and Antigenicity of HIV-1 Env Trimers Resulting from Removal of a Conserved CD4 Binding Site-Proximal Glycan
- PMID: 27489265
- PMCID: PMC5044814
- DOI: 10.1128/JVI.01116-16
Changes in Structure and Antigenicity of HIV-1 Env Trimers Resulting from Removal of a Conserved CD4 Binding Site-Proximal Glycan
Abstract
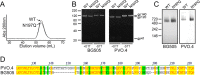
The envelope glycoprotein (Env) is the major target for HIV-1 broadly neutralizing antibodies (bNAbs). One of the mechanisms that HIV has evolved to escape the host's immune response is to mask conserved epitopes on Env with dense glycosylation. Previous studies have shown that the removal of a particular conserved glycan at N197 increases the neutralization sensitivity of the virus to antibodies targeting the CD4 binding site (CD4bs), making it a site of significant interest from the perspective of vaccine design. At present, the structural consequences that result from the removal of the N197 glycan have not been characterized. Using native-like SOSIP trimers, we examine the effects on antigenicity and local structural dynamics resulting from the removal of this glycan. A large increase in the binding of CD4bs and V3-targeting antibodies is observed for the N197Q mutant in trimeric Env, while no changes are observed with monomeric gp120. While the overall structure and thermostability are not altered, a subtle increase in the flexibility of the variable loops at the trimeric interface of adjacent protomers is evident in the N197Q mutant by hydrogen-deuterium exchange mass spectrometry. Structural modeling of the glycan chains suggests that the spatial occupancy of the N197 glycan leads to steric clashes with CD4bs antibodies in the Env trimer but not monomeric gp120. Our results indicate that the removal of the N197 glycan enhances the exposure of relevant bNAb epitopes on Env with a minimal impact on the overall trimeric structure. These findings present a simple modification for enhancing trimeric Env immunogens in vaccines.
Importance: The HIV-1 Env glycoprotein presents a dense patchwork of host cell-derived N-linked glycans. This so-called glycan shield is considered to be a major protective mechanism against immune recognition. While the positions of many N-linked glycans are isolate specific, some are highly conserved and are believed to play key functional roles. In this study, we examine the conserved, CD4 binding site-proximal N197 glycan and demonstrate that its removal both facilitates neutralizing antibody access to the CD4 binding site and modestly impacts the structural dynamics at the trimer crown without drastically altering global Env trimer stability. This indicates that surgical glycosylation site modification may be an effective way of sculpting epitope presentation in Env-based vaccines.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Conserved Role of an N-Linked Glycan on the Surface Antigen of Human Immunodeficiency Virus Type 1 Modulating Virus Sensitivity to Broadly Neutralizing Antibodies against the Receptor and Coreceptor Binding Sites.J Virol. 2015 Oct 28;90(2):829-41. doi: 10.1128/JVI.02321-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26512079 Free PMC article.
-
Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies.J Virol. 2014 Dec;88(24):14002-16. doi: 10.1128/JVI.02614-14. Epub 2014 Sep 24. J Virol. 2014. PMID: 25253346 Free PMC article.
-
Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits.Retrovirology. 2014 May 29;11:41. doi: 10.1186/1742-4690-11-41. Retrovirology. 2014. PMID: 24884783 Free PMC article.
-
Structural Features of Broadly Neutralizing Antibodies and Rational Design of Vaccine.Adv Exp Med Biol. 2018;1075:73-95. doi: 10.1007/978-981-13-0484-2_4. Adv Exp Med Biol. 2018. PMID: 30030790 Review.
-
[Requirements for the Induction of Broadly Neutralizing Antibodies against HIV-1 by Vaccination].Mol Biol (Mosk). 2017 Nov-Dec;51(6):945-957. doi: 10.7868/S0026898417060076. Mol Biol (Mosk). 2017. PMID: 29271959 Review. Russian.
Cited by
-
Evolution of B cell analysis and Env trimer redesign.Immunol Rev. 2017 Jan;275(1):183-202. doi: 10.1111/imr.12515. Immunol Rev. 2017. PMID: 28133805 Free PMC article. Review.
-
An Antigenic Atlas of HIV-1 Escape from Broadly Neutralizing Antibodies Distinguishes Functional and Structural Epitopes.Immunity. 2019 Feb 19;50(2):520-532.e3. doi: 10.1016/j.immuni.2018.12.017. Epub 2019 Jan 29. Immunity. 2019. PMID: 30709739 Free PMC article.
-
Advances in Hydrogen/Deuterium Exchange Mass Spectrometry and the Pursuit of Challenging Biological Systems.Chem Rev. 2022 Apr 27;122(8):7562-7623. doi: 10.1021/acs.chemrev.1c00279. Epub 2021 Sep 7. Chem Rev. 2022. PMID: 34493042 Free PMC article. Review.
-
Structural dynamics reveal isolate-specific differences at neutralization epitopes on HIV Env.iScience. 2022 May 23;25(6):104449. doi: 10.1016/j.isci.2022.104449. eCollection 2022 Jun 17. iScience. 2022. PMID: 35677643 Free PMC article.
-
Engineering well-expressed, V2-immunofocusing HIV-1 envelope glycoprotein membrane trimers for use in heterologous prime-boost vaccine regimens.PLoS Pathog. 2021 Oct 22;17(10):e1009807. doi: 10.1371/journal.ppat.1009807. eCollection 2021 Oct. PLoS Pathog. 2021. PMID: 34679128 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

