The sequential activation of the mitotic microtubule assembly pathways favors bipolar spindle formation
- PMID: 27489339
- PMCID: PMC5042580
- DOI: 10.1091/mbc.E16-05-0322
The sequential activation of the mitotic microtubule assembly pathways favors bipolar spindle formation
Abstract
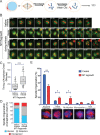
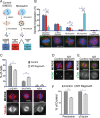
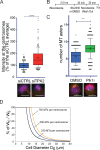
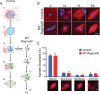
Centrosome maturation is the process by which the duplicated centrosomes recruit pericentriolar components and increase their microtubule nucleation activity before mitosis. The role of this process in cells entering mitosis has been mostly related to the separation of the duplicated centrosomes and thereby to the assembly of a bipolar spindle. However, spindles can form without centrosomes. In fact, all cells, whether they have centrosomes or not, rely on chromatin-driven microtubule assembly to form a spindle. To test whether the sequential activation of these microtubule assembly pathways, defined by centrosome maturation and nuclear envelope breakdown, plays any role in spindle assembly, we combined experiments in tissue culture cells and Xenopus laevis egg extracts with a mathematical model. We found that interfering with the sequential activation of the microtubule assembly pathways compromises bipolar spindle assembly in tissue culture cells but not in X. laevis egg extracts. Our data suggest a novel function for centrosome maturation that determines the contribution of the chromosomal microtubule assembly pathway and favors bipolar spindle formation in most animal cells in which tubulin is in limiting amounts.
© 2016 Cavazza et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
References
-
- Andersen SS. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

