Selecting Cells for Bioartificial Liver Devices and the Importance of a 3D Culture Environment: A Functional Comparison between the HepaRG and C3A Cell Lines
- PMID: 27489500
- PMCID: PMC4971735
- DOI: 10.7150/ijbs.15165
Selecting Cells for Bioartificial Liver Devices and the Importance of a 3D Culture Environment: A Functional Comparison between the HepaRG and C3A Cell Lines
Abstract
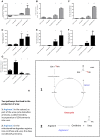
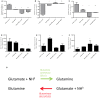
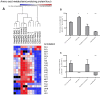
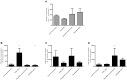
Recently, the first clinical trials on Bioartificial Livers (BALs) loaded with a proliferative human hepatocyte cell source have started. There are two cell lines that are currently in an advanced state of BAL development; HepaRG and HepG2/C3A. In this study we aimed to compare both cell lines on applicability in BALs and to identify possible strategies for further improvement. We tested both cell lines in monolayer- and BAL cultures on growth characteristics, hepatic differentiation, nitrogen-, carbohydrate-, amino acid- and xenobiotic metabolism. Interestingly, both cell lines adapted the hepatocyte phenotype more closely when cultured in BALs; e.g. monolayer cultures produced lactate, while BAL cultures showed diminished lactate production (C3A) or conversion to elimination (HepaRG), and urea cycle activity increased upon BAL culturing in both cell lines. HepaRG-BALs outperformed C3A-BALs on xenobiotic metabolism, ammonia elimination and lactate elimination, while protein synthesis was comparable. In BAL cultures of both cell lines ammonia elimination correlated positively with glutamine production and glutamate consumption, suggesting ammonia elimination was mainly driven by the balance between glutaminase and glutamine synthetase activity. Both cell lines lacked significant urea cycle activity and both required multiple culture weeks before reaching optimal differentiation in BALs. In conclusion, culturing in BALs enhanced hepatic functionality of both cell lines and from these, the HepaRG cells are the most promising proliferative cell source for BAL application.
Keywords: BAL; Bioartificial liver.; C3A; HepaRG; Hepatocytes; Liver support.
Conflict of interest statement
Conflicts of Interest: Robert Chamuleau, is Chief Scientific Officer of the university spin-off company Hep-Art Medical Devices B.V that holds the exclusive licence to the AMC-Bioartificial liver. Ruurdtje Hoekstra was previously employed part-time by Hep-Art Medical Devices B.V.
Valery Shevchenko is an employee of Biopredic International, which currently holds the exclusive license to the HepaRG cell line. The contribution of Biopredic Int. was strictly limited to the technical execution of tests, as proposed by MW and RH. Biopredic Int. had no influence on the content of this article, nor on the decision to publish.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Figures
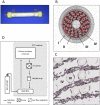
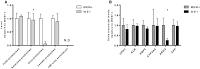
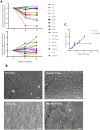
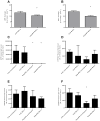
References
-
- Park JK, Lee DH. Bioartificial liver systems: current status and future perspective. Journal of bioscience and bioengineering. 2005;99:311–9. - PubMed
-
- Onions DE, Witt CJ. Xenotransplantation: an overview of microbiological risks and potentials for risk management. Rev Sci Tech. 2000;19:289–301. - PubMed
-
- van Wenum M, Chamuleau RA, van Gulik TM, Siliakus A, Seppen J, Hoekstra R. Bioartificial livers in vitro and in vivo: tailoring biocomponents to the expanding variety of applications. Expert opinion on biological therapy. 2014;14:1745–60. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

