Lollipops in the Clinic: Information Dense Mutation Plots for Precision Medicine
- PMID: 27490490
- PMCID: PMC4973895
- DOI: 10.1371/journal.pone.0160519
Lollipops in the Clinic: Information Dense Mutation Plots for Precision Medicine
Abstract
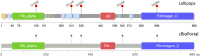
Introduction: Concise visualization is critical to present large amounts of information in a minimal space that can be interpreted quickly. Clinical applications in precision medicine present an important use case due to the time dependent nature of the interpretations, although visualization is increasingly necessary across the life sciences. In this paper we describe the Lollipops software for the presentation of panel or exome sequencing results. Source code and binaries are freely available at https://github.com/pbnjay/lollipops. Although other software and web resources exist to produce lollipop diagrams, these packages are less suited to clinical applications. The demands of precision medicine require the ability to easily fit into a workflow and incorporate external information without manual intervention.
Results: The Lollipops software provides a simple command line interface that only requires an official gene symbol and mutation list making it easily scriptable. External information is integrated using the publicly available Uniprot and Pfam resources. Heuristics are used to select the most informative components and condense them for a concise plot. The output is a flexible Scalable Vector Graphic (SVG) diagram that can be displayed in a web page or graphic illustration tool.
Conclusion: The Lollipops software creates information-dense, publication-quality mutation plots for automated pipelines and high-throughput workflows in precision medicine. The automatic data integration enables clinical data security, and visualization heuristics concisely present knowledge with minimal user configuration.
Conflict of interest statement
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

