Protocols for Molecular Modeling with Rosetta3 and RosettaScripts
- PMID: 27490953
- PMCID: PMC5007558
- DOI: 10.1021/acs.biochem.6b00444
Protocols for Molecular Modeling with Rosetta3 and RosettaScripts
Abstract
Previously, we published an article providing an overview of the Rosetta suite of biomacromolecular modeling software and a series of step-by-step tutorials [Kaufmann, K. W., et al. (2010) Biochemistry 49, 2987-2998]. The overwhelming positive response to this publication we received motivates us to here share the next iteration of these tutorials that feature de novo folding, comparative modeling, loop construction, protein docking, small molecule docking, and protein design. This updated and expanded set of tutorials is needed, as since 2010 Rosetta has been fully redesigned into an object-oriented protein modeling program Rosetta3. Notable improvements include a substantially improved energy function, an XML-like language termed "RosettaScripts" for flexibly specifying modeling task, new analysis tools, the addition of the TopologyBroker to control conformational sampling, and support for multiple templates in comparative modeling. Rosetta's ability to model systems with symmetric proteins, membrane proteins, noncanonical amino acids, and RNA has also been greatly expanded and improved.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
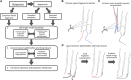


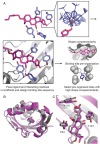
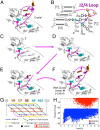
References
-
- Raman S.; Vernon R.; Thompson J.; Tyka M.; Sadreyev R.; Pei J.; Kim D.; Kellogg E.; DiMaio F.; Lange O.; Kinch L.; Sheffler W.; Kim B. H.; Das R.; Grishin N. V.; Baker D. (2009) Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins: Struct., Funct., Genet. 77 (Suppl. 9), 89–99. 10.1002/prot.22540. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

