Multiplex 16S rRNA-derived geno-biochip for detection of 16 bacterial pathogens from contaminated foods
- PMID: 27492058
- PMCID: PMC5132059
- DOI: 10.1002/biot.201600043
Multiplex 16S rRNA-derived geno-biochip for detection of 16 bacterial pathogens from contaminated foods
Abstract
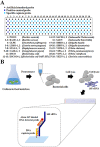
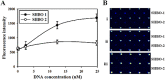
Foodborne diseases caused by various pathogenic bacteria occur worldwide. To prevent foodborne diseases and minimize their impacts, it is important to inspect contaminated foods and specifically detect many types of pathogenic bacteria. Several DNA oligonucleotide biochips based on 16S rRNA have been investigated to detect bacteria; however, a mode of detection that can be used to detect diverse pathogenic strains and to examine the safety of food matrixes is still needed. In the present work, a 16S rRNA gene-derived geno-biochip detection system was developed after screening DNA oligonucleotide specific capture probes, and it was validated for multiple detection of 16 pathogenic strains that frequently occur with a signature pattern. rRNAs were also used as detection targets directly obtained from cell lysates without any purification and amplification steps in the bacterial cells separated from 8 food matrixes by simple pretreatments. Thus, the developed 16S rRNA-derived geno-biochip can be successfully used for the rapid and multiple detection of the 16 pathogenic bacteria frequently isolated from contaminated foods that are important for food safety.
Keywords: Direct RNA-detection; Food matrix; Foodborne pathogen; Multiple detection; Oligonucleotide microarray.
© 2016 The Authors. Biotechnology Journal published by WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


Similar articles
-
Development and application of an oligonucleotide microarray for the detection of food-borne bacterial pathogens.Appl Microbiol Biotechnol. 2007 Aug;76(1):225-33. doi: 10.1007/s00253-007-0993-x. Epub 2007 May 10. Appl Microbiol Biotechnol. 2007. PMID: 17492283
-
Multiple detection of food-borne pathogenic bacteria using a novel 16S rDNA-based oligonucleotide signature chip.Biosens Bioelectron. 2007 Jan 15;22(6):845-53. doi: 10.1016/j.bios.2006.03.005. Epub 2006 Apr 18. Biosens Bioelectron. 2007. PMID: 16621503
-
Specific multiplex analysis of pathogens using a direct 16S rRNA hybridization in microarray system.Anal Chem. 2012 Jun 5;84(11):4873-9. doi: 10.1021/ac300476k. Epub 2012 May 11. Anal Chem. 2012. PMID: 22551354
-
Development and application of new nucleic acid-based technologies for microbial community analyses in foods.Int J Food Microbiol. 2002 Sep 15;78(1-2):171-80. doi: 10.1016/s0168-1605(02)00236-2. Int J Food Microbiol. 2002. PMID: 12222632 Review.
-
[16S rRNA gene sequence analysis for bacterial identification in the clinical laboratory].Rinsho Byori. 2013 Dec;61(12):1107-15. Rinsho Byori. 2013. PMID: 24605544 Review. Japanese.
Cited by
-
Simultaneous Detection of Five Foodborne Pathogens Using a Mini Automatic Nucleic Acid Extractor Combined with Recombinase Polymerase Amplification and Lateral Flow Immunoassay.Microorganisms. 2022 Jul 5;10(7):1352. doi: 10.3390/microorganisms10071352. Microorganisms. 2022. PMID: 35889071 Free PMC article.
References
-
- Korean Ministry of Food and Drug Safety, The Statistic System For Foodborne Disease. 2015.
-
- Linscott, A. J. , Food‐Borne Illnesses. Clin. Microbiol. Newsl. 2011, 33, 41–45.
-
- Aytac, S. , Taban, B. , Food‐borne microbial diseases and control: Food‐borne infections and intoxications, in: Malik A., Erginkaya Z., Ahmad S., Erten H. (Eds.), Food Processing: Strategies for Quality Assessment, Springer; New York: 2014, pp. 191–224.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

