PcircRNA_finder: a software for circRNA prediction in plants
- PMID: 27493192
- PMCID: PMC5181569
- DOI: 10.1093/bioinformatics/btw496
PcircRNA_finder: a software for circRNA prediction in plants
Abstract
Motivation: Recent studies reveal an important role of non-coding circular RNA (circRNA) in the control of cellular processes. Because of differences in the organization of plant and mammal genomes, the sensitivity and accuracy of circRNA prediction programs using algorithms developed for animals and humans perform poorly for plants.
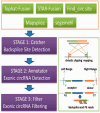
Results: A circRNA prediction software for plants (termed PcircRNA_finder) was developed that is more sensitive in detecting circRNAs than other frequently used programs (such as find_circ and CIRCexplorer), Based on analysis of simulated and real rRNA-/RNAase R RNA-Seq data from Arabidopsis thaliana and rice PcircRNA_finder provides a more comprehensive sensitive, precise prediction method for plants circRNAs.
Availability and implementation: http://ibi.zju.edu.cn/bioinplant/tools/manual.htm CONTACT: fanlj@zju.edu.cnSupplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures
References
-
- Hansen T.B. et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources