Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases
- PMID: 27493650
- PMCID: PMC4955380
- DOI: 10.3389/fpls.2016.01045
Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases
Abstract
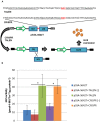
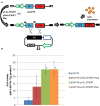
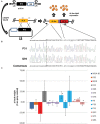
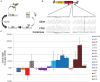
Genome editing using sequence-specific nucleases (SSNs) is rapidly being developed for genetic engineering in crop species. The utilization of zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated systems (CRISPR/Cas) for inducing double-strand breaks facilitates targeting of virtually any sequence for modification. Targeted mutagenesis via non-homologous end-joining (NHEJ) has been demonstrated extensively as being the preferred DNA repair pathway in plants. However, gene targeting via homologous recombination (HR) remains more elusive but could be a powerful tool for directed DNA repair. To overcome barriers associated with gene targeting, a geminivirus replicon (GVR) was used to deliver SSNs targeting the potato ACETOLACTATE SYNTHASE1 (ALS1) gene and repair templates designed to incorporate herbicide-inhibiting point mutations within the ALS1 locus. Transformed events modified with GVRs held point mutations that were capable of supporting a reduced herbicide susceptibility phenotype, while events transformed with conventional T-DNAs held no detectable mutations and were similar to wild-type. Regeneration of transformed events improved detection of point mutations that supported a stronger reduced herbicide susceptibility phenotype. These results demonstrate the use of geminiviruses for delivering genome editing reagents in plant species, and a novel approach to gene targeting in a vegetatively propagated species.
Keywords: CRISPR/Cas; TALEN; ZFN; acetolactate synthase; bean yellow dwarf virus; gene replacement; gene targeting; homologous recombination.
Figures

References
-
- Ascencio-Ibáñez J. T., Sozzani R., Lee T.-J., Chu T.-M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 436–454. 10.1104/pp.108.121038 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

