MLST and Whole-Genome-Based Population Analysis of Cryptococcus gattii VGIII Links Clinical, Veterinary and Environmental Strains, and Reveals Divergent Serotype Specific Sub-populations and Distant Ancestors
- PMID: 27494185
- PMCID: PMC4975453
- DOI: 10.1371/journal.pntd.0004861
MLST and Whole-Genome-Based Population Analysis of Cryptococcus gattii VGIII Links Clinical, Veterinary and Environmental Strains, and Reveals Divergent Serotype Specific Sub-populations and Distant Ancestors
Abstract
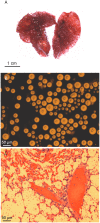
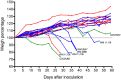
The emerging pathogen Cryptococcus gattii causes life-threatening disease in immunocompetent and immunocompromised hosts. Of the four major molecular types (VGI-VGIV), the molecular type VGIII has recently emerged as cause of disease in otherwise healthy individuals, prompting a need to investigate its population genetic structure to understand if there are potential genotype-dependent characteristics in its epidemiology, environmental niche(s), host range and clinical features of disease. Multilocus sequence typing (MLST) of 122 clinical, environmental and veterinary C. gattii VGIII isolates from Australia, Colombia, Guatemala, Mexico, New Zealand, Paraguay, USA and Venezuela, and whole genome sequencing (WGS) of 60 isolates representing all established MLST types identified four divergent sub-populations. The majority of the isolates belong to two main clades, corresponding either to serotype B or C, indicating an ongoing species evolution. Both major clades included clinical, environmental and veterinary isolates. The C. gattii VGIII population was genetically highly diverse, with minor differences between countries, isolation source, serotype and mating type. Little to no recombination was found between the two major groups, serotype B and C, at the whole and mitochondrial genome level. C. gattii VGIII is widespread in the Americas, with sporadic cases occurring elsewhere, WGS revealed Mexico and USA as a likely origin of the serotype B VGIII population and Colombia as a possible origin of the serotype C VGIII population. Serotype B isolates are more virulent than serotype C isolates in a murine model of infection, causing predominantly pulmonary cryptococcosis. No specific link between genotype and virulence was observed. Antifungal susceptibility testing against six antifungal drugs revealed that serotype B isolates are more susceptible to azoles than serotype C isolates, highlighting the importance of strain typing to guide effective treatment to improve the disease outcome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Kwon-Chung KJ, Boekhout T, Wickes BL, Fell JW. Systematics of Genus Cryptococcus and its type species C. neoformans In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Cryptococcus: From Human Pathogen to Model Yeast. Washington, DC: ASM Press; 2011; pp. 3–15.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

