Comparative genomics and transcriptomics of Pichia pastoris
- PMID: 27495311
- PMCID: PMC4974788
- DOI: 10.1186/s12864-016-2876-y
Comparative genomics and transcriptomics of Pichia pastoris
Erratum in
-
Erratum to: Comparative genomics and transcriptomics of Pichia pastoris.BMC Genomics. 2016 Sep 28;17(1):762. doi: 10.1186/s12864-016-3109-0. BMC Genomics. 2016. PMID: 27681084 Free PMC article. No abstract available.
Abstract
Background: Pichia pastoris has emerged as an important alternative host for producing recombinant biopharmaceuticals, owing to its high cultivation density, low host cell protein burden, and the development of strains with humanized glycosylation. Despite its demonstrated utility, relatively little strain engineering has been performed to improve Pichia, due in part to the limited number and inconsistent frameworks of reported genomes and transcriptomes. Furthermore, the co-mingling of genomic, transcriptomic and fermentation data collected about Komagataella pastoris and Komagataella phaffii, the two strains co-branded as Pichia, has generated confusion about host performance for these genetically distinct species. Generation of comparative high-quality genomes and transcriptomes will enable meaningful comparisons between the organisms, and potentially inform distinct biotechnological utilies for each species.
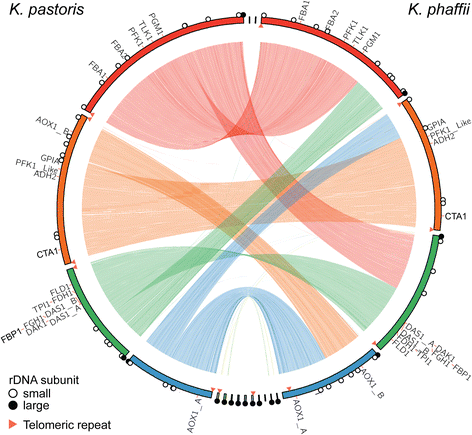
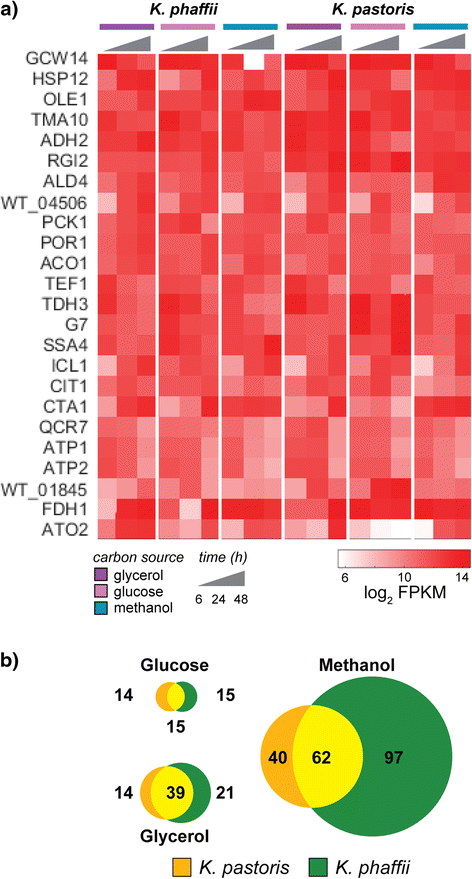
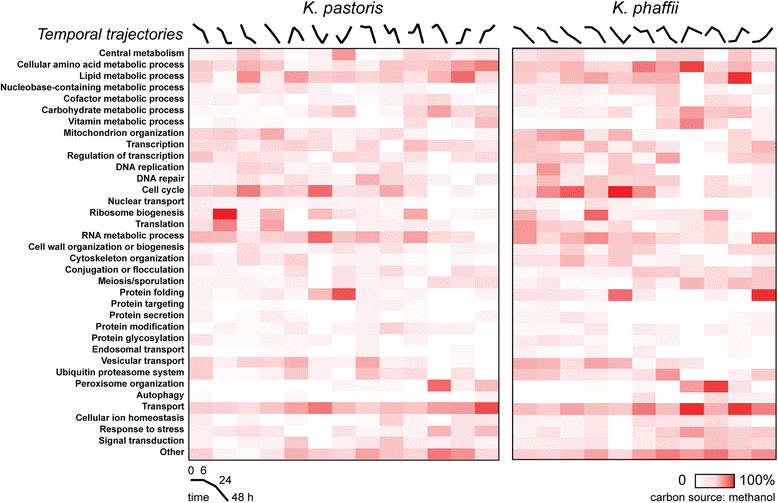
Results: Here, we present a comprehensive and standardized comparative analysis of the genomic features of the three most commonly used strains comprising the tradename Pichia: K. pastoris wild-type, K. phaffii wild-type, and K. phaffii GS115. We used a combination of long-read (PacBio) and short-read (Illumina) sequencing technologies to achieve over 1000X coverage of each genome. Construction of individual genomes was then performed using as few as seven individual contigs to create gap-free assemblies. We found substantial syntenic rearrangements between the species and characterized a linear plasmid present in K. phaffii. Comparative analyses between K. phaffii genomes enabled the characterization of the mutational landscape of the GS115 strain. We identified and examined 35 non-synonomous coding mutations present in GS115, many of which are likely to impact strain performance. Additionally, we investigated transcriptomic profiles of gene expression for both species during cultivation on various carbon sources. We observed that the most highly transcribed genes in both organisms were consistently highly expressed in all three carbon sources examined. We also observed selective expression of certain genes in each carbon source, including many sequences not previously reported as promoters for expression of heterologous proteins in yeasts.
Conclusions: Our studies establish a foundation for understanding critical relationships between genome structure, cultivation conditions and gene expression. The resources we report here will inform and facilitate rational, organism-wide strain engineering for improved utility as a host for protein production.
Keywords: Cultivation dependent expression; Gene Set Enrichment Analysis (GSEA); Komagataella pastoris; Komagataella phaffii; Pichia pastoris; RNA-Seq; Secretome; Self-Organizing Maps (SOMs); Transcriptome.
Figures



References
-
- Corchero JL, Gasser B, Resina D, Smith W, Parrilli E, Vazquez F, Abasolo I, Giuliani M, Jantti J, Ferrer P, et al. Unconventional microbial systems for the cost-efficient production of high-quality protein therapeutics. Biotechnol Adv. 2013;31(2):140–53. doi: 10.1016/j.biotechadv.2012.09.001. - DOI - PubMed
-
- Love JC, Love KR, Barone PW. Enabling global access to high-quality biopharmaceuticals. Curr Opin Chem Eng. 2013;2(4):383–90. doi: 10.1016/j.coche.2013.09.002. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

