The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans
- PMID: 27496166
- PMCID: PMC4975890
- DOI: 10.1186/s12864-016-2837-5
The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans
Abstract
Background: The heat shock response, induced by cytoplasmic proteotoxic stress, is one of the most highly conserved transcriptional responses. This response, driven by the heat shock transcription factor HSF1, restores proteostasis through the induction of molecular chaperones and other genes. In addition to stress-dependent functions, HSF1 has also been implicated in various stress-independent functions. In C. elegans, the HSF1 homolog HSF-1 is an essential protein that is required to mount a stress-dependent response, as well as to coordinate various stress-independent processes including development, metabolism, and the regulation of lifespan. In this work, we have performed RNA-sequencing for C. elegans cultured in the presence and absence of hsf-1 RNAi followed by treatment with or without heat shock. This experimental design thus allows for the determination of both heat shock-dependent and -independent biological targets of HSF-1 on a genome-wide level.
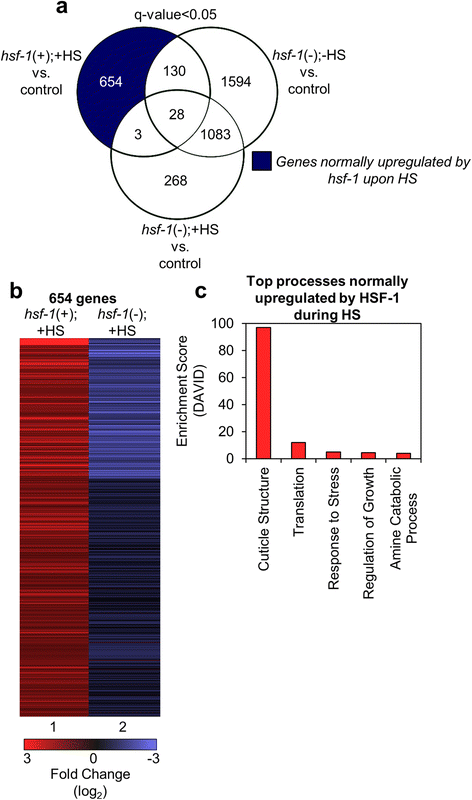
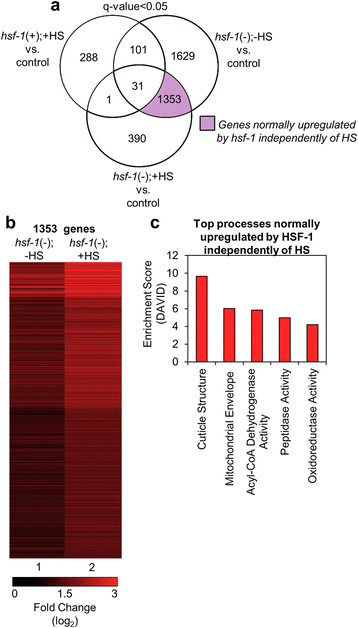
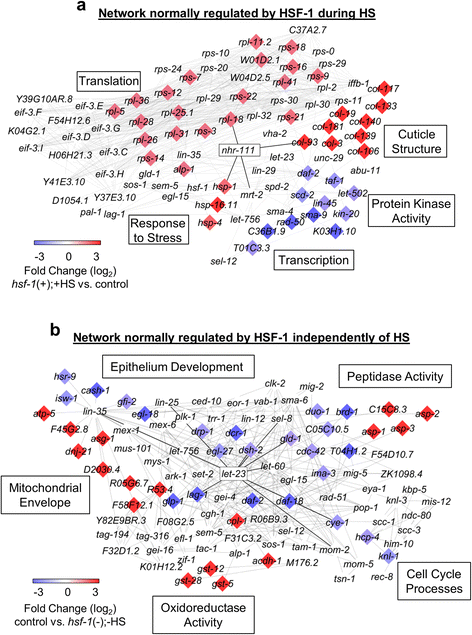
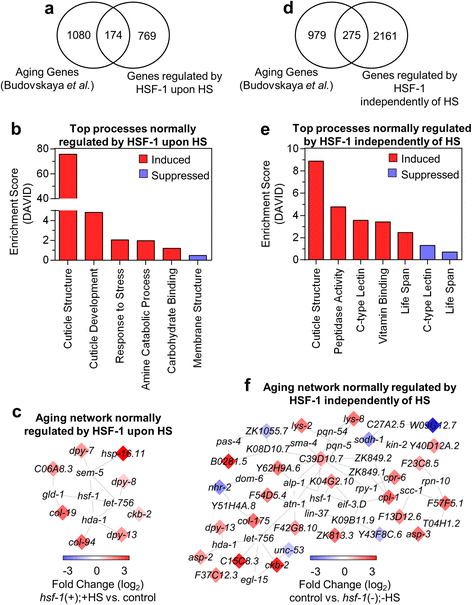
Results: Our results confirm that C. elegans HSF-1 can regulate gene expression in both a stress-dependent and -independent fashion. Almost all genes regulated by HS require HSF-1, reinforcing the central role of this transcription factor in the response to heat stress. As expected, major categories of HSF-1-regulated genes include cytoprotection, development, metabolism, and aging. Within both the heat stress-dependent and -independent gene groups, significant numbers of genes are upregulated as well as downregulated, demonstrating that HSF-1 can both activate and repress gene expression either directly or indirectly. Surprisingly, the cellular process most highly regulated by HSF-1, both with and without heat stress, is cuticle structure. Via network analyses, we identify a nuclear hormone receptor as a common link between genes that are regulated by HSF-1 in a HS-dependent manner, and an epidermal growth factor receptor as a common link between genes that are regulated by HSF-1 in a HS-independent manner. HSF-1 therefore coordinates various physiological processes in C. elegans, and HSF-1 activity may be coordinated across tissues by nuclear hormone receptor and epidermal growth factor receptor signaling.
Conclusion: This work provides genome-wide HSF-1 regulatory networks in C. elegans that are both heat stress-dependent and -independent. We show that HSF-1 is responsible for regulating many genes outside of classical heat stress-responsive genes, including genes involved in development, metabolism, and aging. The findings that a nuclear hormone receptor may coordinate the HS-induced HSF-1 transcriptional response, while an epidermal growth factor receptor may coordinate the HS-independent response, indicate that these factors could promote cell non-autonomous signaling that occurs through HSF-1. Finally, this work highlights the genes involved in cuticle structure as important HSF-1 targets that may play roles in promoting both cytoprotection as well as longevity.
Keywords: C. elegans; HSF-1; Heat shock response; RNA-seq; Stress; Transcript analysis.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

