Using cell deformation and motion to predict forces and collective behavior in morphogenesis
- PMID: 27496334
- PMCID: PMC5290285
- DOI: 10.1016/j.semcdb.2016.07.029
Using cell deformation and motion to predict forces and collective behavior in morphogenesis
Abstract
In multi-cellular organisms, morphogenesis translates processes at the cellular scale into tissue deformation at the scale of organs and organisms. To understand how biochemical signaling regulates tissue form and function, we must understand the mechanical forces that shape cells and tissues. Recent progress in developing mechanical models for tissues has led to quantitative predictions for how cell shape changes and polarized cell motility generate forces and collective behavior on the tissue scale. In particular, much insight has been gained by thinking about biological tissues as physical materials composed of cells. Here we review these advances and discuss how they might help shape future experiments in developmental biology.
Keywords: Collective motion; Deformation; Epithelium; Jamming; Morphogenesis; Tissue mechanics.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
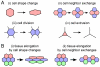
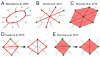



References
-
- Fung YC. Biomechanics: Mechanical Properties of Living Tissues. Second Edition Springer; 1993.
-
- Ranft J, Basan M, Elgeti J, Joanny J-F, Prost J, Julicher F. Fluidization of tissues by cell division and apoptosis. Proceedings of the National Academy of Sciences. 2010;107(49):20863–20868. doi:10.1073/pnas.1011086107. URL http://www.pnas.org/cgi/doi/10.1073/pnas.1011086107. - DOI - PMC - PubMed
-
- Puliafito A, Hufnagel L, Neveu P, Streichan S, Sigal A, Fygenson DK, Shraiman BI. Collective and single cell behavior in epithelial contact inhibition. Proceedings of the National Academy of Sciences. 2012;109(3):739–744. doi:10.1073/pnas.1007809109. URL http://www.pnas.org/cgi/doi/10.1073/pnas.1007809109. - DOI - PMC - PubMed
-
- von Dassow M, Davidson L. a. Physics and the canalization of morphogenesis: a grand challenge in organismal biology. Physical Biology. 2011;8(4):045002. doi:10.1088/1478-3975/8/4/045002. URL http://stacks.iop.org/1478-3975/8/i=4/a=045002?key=crossref.502250c37be0.... - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

