Integrating metabolomics and transcriptomics data to discover a biocatalyst that can generate the amine precursors for alkamide biosynthesis
- PMID: 27497272
- PMCID: PMC5195896
- DOI: 10.1111/tpj.13295
Integrating metabolomics and transcriptomics data to discover a biocatalyst that can generate the amine precursors for alkamide biosynthesis
Abstract
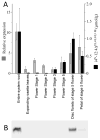
The Echinacea genus is exemplary of over 30 plant families that produce a set of bioactive amides, called alkamides. The Echinacea alkamides may be assembled from two distinct moieties, a branched-chain amine that is acylated with a novel polyunsaturated fatty acid. In this study we identified the potential enzymological source of the amine moiety as a pyridoxal phosphate-dependent decarboxylating enzyme that uses branched-chain amino acids as substrate. This identification was based on a correlative analysis of the transcriptomes and metabolomes of 36 different E. purpurea tissues and organs, which expressed distinct alkamide profiles. Although no correlation was found between the accumulation patterns of the alkamides and their putative metabolic precursors (i.e., fatty acids and branched-chain amino acids), isotope labeling analyses supported the transformation of valine and isoleucine to isobutylamine and 2-methylbutylamine as reactions of alkamide biosynthesis. Sequence homology identified the pyridoxal phosphate-dependent decarboxylase-like proteins in the translated proteome of E. purpurea. These sequences were prioritized for direct characterization by correlating their transcript levels with alkamide accumulation patterns in different organs and tissues, and this multi-pronged approach led to the identification and characterization of a branched-chain amino acid decarboxylase, which would appear to be responsible for generating the amine moieties of naturally occurring alkamides.
Keywords: Echinacea purpurea; alkamides; amines; fatty acids; metabolomics; specialized metabolism; transcriptomics.
© 2016 The Authors The Plant Journal © 2016 John Wiley & Sons Ltd.
Figures





References
-
- Bauer R, Foster S. Analysis of alkamides and caffeic acid derivatives from Echinacea simulata and E. paradoxa roots. Planta Med. 1991;57:447–449. - PubMed
-
- Bauer R, Khan IA, Wagner H. TLC and HPLC Analysis of Echinacea pallida and E. angustifolia Roots1. Planta Med. 1988a;54:426–430. - PubMed
-
- Bauer R, Remiger P. TLC and HPLC Analysis of Alkamides in Echinacea Drugs1,2. Planta Med. 1989;55:367–371. - PubMed
-
- Bauer R, Remiger P, Wagner H. Alkamides from the roots of Echinacea purpurea. Phytochemistry. 1988b;27:2339–2342.
-
- Bauer R, Remiger P, Wagner H. Alkamides from the roots of Echinacea angustifolia. Phytochemistry. 1989;28:505–508.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

