NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation
- PMID: 27497299
- PMCID: PMC5048346
- DOI: 10.15252/embj.201694885
NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation
Abstract
Mitochondrial gene expression uses a non-universal genetic code in mammals. Besides reading the conventional AUG codon, mitochondrial (mt-)tRNAMet mediates incorporation of methionine on AUA and AUU codons during translation initiation and on AUA codons during elongation. We show that the RNA methyltransferase NSUN3 localises to mitochondria and interacts with mt-tRNAMet to methylate cytosine 34 (C34) at the wobble position. NSUN3 specifically recognises the anticodon stem loop (ASL) of the tRNA, explaining why a mutation that compromises ASL basepairing leads to disease. We further identify ALKBH1/ABH1 as the dioxygenase responsible for oxidising m5C34 of mt-tRNAMet to generate an f5C34 modification. In vitro codon recognition studies with mitochondrial translation factors reveal preferential utilisation of m5C34 mt-tRNAMet in initiation. Depletion of either NSUN3 or ABH1 strongly affects mitochondrial translation in human cells, implying that modifications generated by both enzymes are necessary for mt-tRNAMet function. Together, our data reveal how modifications in mt-tRNAMet are generated by the sequential action of NSUN3 and ABH1, allowing the single mitochondrial tRNAMet to recognise the different codons encoding methionine.
Keywords: ABH1; NSUN3; RNA modification; mitochondria; translation.
© 2016 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures

The localisation of NSUN3 was analysed in HEK293 cells stably expressing NSUN3‐GFP. NSUN3‐GFP (green) and staining with Mitotracker (red) are shown separately and in an overlay with DAPI to indicate nuclei. The scale bar represents 5 μm.
To analyse submitochondrial localisation of NSUN3, human mitochondria were isolated and either left untreated, swollen in hypotonic buffer (Mitoplasts) or disrupted by sonication (Sonic.) before treatment with different amounts of proteinase K (PK) where indicated, followed by SDS–PAGE and Western blotting using antibodies against human TIM44, TIM23, TOM70 or FLAG‐tagged NSUN3. Note that TIM44 extends into the matrix, while the N‐terminus of TIM23 localises to the intermembrane space and TOM70 is largely exposed on the mitochondrial surface. The asterisk indicates a cross‐reaction of the TOM70 antibody.
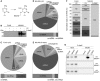
- A
Structure of 5‐azacytidine and formation of a covalent RNA methyltransferase adduct.
- B
HEK293 cells expressing NSUN3‐HisPrcFLAG (NSUN3) or the HisPrcFLAG tag alone (FLAG) were either not cross‐linked (−), UV cross‐linked (UV) or treated with 5‐azacytidine (5‐AzaC). The protein–RNA complexes were affinity purified and the bound RNA was trimmed, end‐labelled with 32P phosphate and ligated to linkers. Protein–RNA complexes were separated by SDS–PAGE, transferred to nitrocellulose and exposed to an X‐ray film.
- C–E
The UV or 5‐AzaC cross‐linking and analysis of cDNA (CRAC) experiments with NSUN3‐HisPrcFLAG (D, E) or the FLAG control (C) samples were treated as described in (B). The RNA was isolated from the nitrocellulose membrane‐bound protein–RNA complexes and converted into cDNA for sequence library production and Illumina deep sequencing. Pie charts present different RNA classes and the relative distribution of Illumina sequence reads that were obtained after mapping of the reads on the human genome. Bar graphs below indicate the distribution of mitochondrial (mt‐)tRNA, mt‐rRNA and mt‐mRNA sequence reads among the reads mapped to the mitochondrial genome. Abbreviations: tRNA, transfer RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; rRNA, ribosomal RNA; mtRNA, mitochondrial‐encoded RNA; miscRNA, miscellaneous RNA; miRNA, microRNA; lncRNA, long non‐coding RNA.
- F
Relative distribution of mitochondrial tRNA sequence reads obtained from the CRAC experiments using UV or 5‐AzaC cross‐linking with cells expressing the NSUN3‐HisPrcFLAG (NSUN3) protein or control cells (FLAG). Only mt‐tRNAs that were represented by more than 5% of all mt‐tRNAs reads are labelled.
- G
5‐AzaC cross‐linking was performed and RNA associated with wild‐type NSUN3, the catalytically inactive NSUN3 mutant (C265A) or the FLAG tag alone was isolated as described in (B). The RNA was analysed by Northern blot using probes against the mt‐tRNAMet, mt‐tRNAPro and mt‐tRNAGlu. Inputs (0.1%) are shown on the left and eluates (50%) on the right.

- A, B
The UV or 5‐AzaC cross‐linking and analysis of cDNA (CRAC) experiments with NSUN3‐HisPrcFLAG or FLAG control cells were performed as described for Fig 2. (A) The percentages of the Illumina sequence reads mapped to individual classes of RNA are given graphically for each sample. Abbreviations: tRNA, transfer RNA; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; rRNA, ribosomal RNA; mtRNA, mitochondrial‐encoded RNA; miscRNA, miscellaneous RNA; miRNA, microRNA; lncRNA, long non‐coding RNA. (B) The relative distribution of cytoplasmic tRNA sequence reads obtained from the CRAC experiments is shown. Only tRNAs that were represented by more than 5% of all cytoplasmic tRNA reads are labelled.
- C
In vitro methylation reactions were performed using recombinant His14‐MBP‐NSUN3 (NSUN3) or the catalytically inactive mutant His14‐MBP‐NSUN3‐C265A (C265A), [3H‐methyl]‐labelled S‐adenosylmethionine as a methyl group donor and in vitro‐transcribed mitochondrial mt‐tRNAMet, cytoplasmic tRNAi Met and tRNAe Met. The RNA was then separated on a denaturing polyacrylamide gel, stained with ethidium bromide (EtBr) to indicate inputs and exposed to an X‐ray film to analyse methylation (3H‐Me).
- D
5‐AzaC cross‐linking was performed and RNA‐associated with wild‐type NSUN3, the catalytic NSUN3 mutant (C265A) or the FLAG tag alone was isolated as described in (A). The RNA was isolated from the purified protein–RNA complexes and analysis by Northern blot using probes against the mt‐tRNAMet, mt‐tRNAi Met and mt‐tRNAe Met. Inputs are shown on the left and eluates on the right. The mt‐tRNAMet panel is identical to that shown in Fig 2G.
- E
The nucleotide sequences of the anticodon stem loops of mt‐tRNAMet (left) and tRNAi Met (right) are shown.
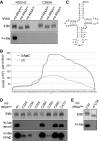
In vitro methylation reactions were performed using recombinant His14‐MBP‐NSUN3 (NSUN3) or the catalytically inactive mutant His14‐MBP‐NSUN3‐C265A (C265A), 3H‐labelled S‐adenosylmethionine as a methyl group donor and in vitro‐transcribed mt‐tRNAMet, mt‐tRNAPro and mt‐tRNAGlu. The RNA was then separated on a denaturing polyacrylamide gel, stained with ethidium bromide (EtBr) to indicate inputs and exposed to an X‐ray film to analyse methylation (3H‐Me).
The distribution of Illumina sequence reads along the mt‐tRNAMet sequence obtained from CRAC experiments with NSUN3 after UV (light grey) or 5‐AzaC cross‐linking (dark grey) is given as reads per million mapped reads. The position of the anticodon is indicated by a bar.
Cloverleaf scheme of the mt‐tRNAMet sequence. Nucleosides that were exchanged in the mutational analysis shown in the following panels are marked with arrows, and the nucleotide positions in the tRNA are given.
In vitro methylation assays were performed as described in (A) with His14‐MBP‐NSUN3 and in vitro‐transcribed wild‐type mt‐tRNAMet and cytidine mutants of the anticodon stem and loop region indicated in (C). Two exposure times of X‐ray films are shown 16 h (short) and 3 days (long).
In vitro methylation assay of in vitro‐transcribed mt‐tRNAMet and chemically synthesised mt‐tRNAMet containing an m5C modification at the wobble position. The experiment and analysis were performed as described in (A).

Scheme showing the mutations introduced in the anticodon stem loop (ASL) of mt‐tRNAMet for analysing tRNA substrate recognition by NSUN3. ASL mutants included G:U basepairs (G:U bp) to affect the stability of basepairing and sequence of the stem or mutants were generated by swapping G:C basepairs (G/C swap), leading to changes in sequence without affecting basepairing stability.
In vitro methylation assays were performed using [3H‐methyl]‐labelled S‐adenosylmethionine, the in vitro transcripts of the mt‐tRNAMet mutants described in (A) and recombinant His14‐MBP‐NSUN3. RNA was then separated on a denaturing polyacrylamide gel, stained with ethidium bromide (EtBr), dried and exposed to an X‐ray film to detect methylated transcripts (3H‐Me).
In vitro methylation assay using chemically synthesised ASL. The experiment and analysis were performed as described in (B).
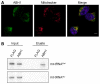
The localisation of ABH1 was analysed by immunofluorescence in HEK293 cells. ABH1 (green) localisation and mitochondria stained with Mitotracker (red) are shown separately and in an overlay with DAPI to indicate nuclei. The scale bar represents 5 μm.
HEK293 cells expressing ABH1‐HisPrcFLAG (ABH1) or the HisPrcFLAG tag alone (FLAG) were UV cross‐linked (UV), and protein–RNA complexes were affinity purified. Co‐precipitated RNA was isolated and analysed by Northern blot using probes against mt‐tRNAMet and mt‐tRNAGlu. Inputs (0.1%) are shown on the left and eluates (50%) on the right.


In vitro‐transcribed mt‐tRNAMet was methylated at C34 using recombinant NSUN3 and 3H‐labelled S‐adenosylmethionine as a methyl group donor. Radiolabelled mt‐tRNAMet was re‐extracted and then subjected to oxidation assays without protein (−), with maltose binding protein (MBP), with the dioxygenase FTO or using wild‐type (ABH1) or mutant (R338A, D233A) His14‐MBP‐ABH1. Besides ABH1 controls lacking α‐ketoglutarate (αKG) or Fe2+ ions, all samples contained α‐ketoglutarate and Fe2+ ions. After oxidation, RNA was precipitated and the tritium released upon oxidation of radiolabelled mt‐tRNAMet was quantified in the supernatant. Counts per minute (CPM) are shown for experiments performed in triplicate with error bars indicating ± SD (upper panel). Pelleted RNA was separated on a denaturing polyacrylamide gel and exposed to an X‐ray film to analyse the tritium retained (3H‐Me).
Synthetic anticodon stem loop (ASL) was radioactively labelled and subjected to oxidation assays that were performed and analysed as described in (A) using no protein (−), MBP or wild‐type His14‐MBP‐ABH1 (ABH1). Experiments were performed in triplicate with error bars indicating ± SD.
Anion exchange HPLC analysis was performed on synthetic m5C‐containing ASL (20 nt) before and after oxidation by ABH1. The small shift in retention time indicates formation of f5C‐modified RNA. The ABH1 oxidation product was then treated with NaBH4 to generate hm5C‐modified RNA. All three samples were analysed by ESI‐MS, and the molecular weight (m.w.) is indicated on the HPLC trace. Only the f5C‐containing RNA was labelled efficiently with 1‐ethyl‐2,3,3‐trimethylindoleninium‐5‐sulphonate (TMI).
The different retention times of m5C‐, hm5C‐ and f5C‐modified ASL RNA were confirmed by co‐injection of samples shown in (C). HPLC was performed as in (C).
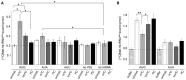
MTIF2‐dependent reading of initiation codons AUG, AUA or AUU in the P site of the ribosome by unmodified (unmod.) or C34‐modified [14C]Met‐tRNAMet. Binding was determined by nitrocellulose filtration, and [14C]Met‐tRNAMet retrieved on the membrane was quantified by scintillation counting. Binding in the absence of ribosomes (no 70S) or mRNA (no mRNA) served as controls. Data from three independent experiments are presented with error bars indicating ± SEM. The statistical significance of the results was analysed by t‐test and is indicated by the asterisks in the graph (*P < 0.05).
TUFM‐dependent recognition of A site codons during elongation. Data from three independent experiments are presented with error bars indicating ± SEM and statistical analysis as in (A) (*P < 0.05, **P < 0.01).

HeLa cells were transfected with two different siRNAs against NSUN3 (siNSUN3_1, siNSUN3_2), ABH1 (siABH1_1, siABH1_2) or with non‐target (siNT) siRNA, and the knock‐down efficiency was analysed by quantitative PCR. The relative abundance of the NSUN3 or ABH1 mRNA was normalised to GAPDH levels. Data are presented as mean ± SD.
Chemically synthesised f5C modified mt‐tRNAMet and total RNA from wild‐type (WT) cells or those transfected with siRNAs as in (A) were treated with TMI to specifically label f5C residues. Primer extension, using a radiolabelled antisense primer, was performed under limited dNTP conditions. Products were separated on a denaturing polyacrylamide gel alongside a sequencing ladder, and RNAs were detected using a phosphorimager.
Primer extension reactions were performed on total RNA from cells transfected with siRNAs as described in (B). Stops corresponding to position C34 in mt‐tRNAMet were quantified in three independent experiments, and results are shown graphically as mean ± SD.
RNA from wild‐type HeLa cells and cells treated with siRNAs against NSUN3 or ABH1 (as in A) was either first reduced with NaBH4 or directly treated with bisulphite. After deamination and desulphonation, mt‐tRNAMet RNAs were reverse transcribed, amplified, cloned and sequenced. The proportions of thymine (grey) generated by bisulphite conversion or non‐converted cytosine (black) at position 34 of mt‐tRNAMet are shown. Note that for sequences from non‐reduced samples, thymine can also originate from unmodified or f5C‐containing mt‐tRNAMet, while in reduced samples, it originates from unmodified cytosine.

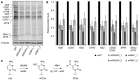
HeLa cells were treated with non‐target siRNAs (siNT) or those targeting NSUN3 (siNSUN3_1 or siNSUN3_2) or ABH1 (siABH1_1 or siABH1_2) for 72 h before labelling of mitochondrial translation products with [35S]methionine. Protein samples were separated by SDS–PAGE then transferred to a membrane. Labelled proteins were detected using a phosphorimager, and the levels of tubulin were determined by Western blotting using an antibody against the endogenous protein for normalisation.
Mitochondrially translated proteins that could be clearly detected were quantified in three independent experiments, and the results are shown graphically as mean ± SD.
Overview of the modification pathway of C34 in mt‐tRNAMet. NSUN3 introduces an m5C methylation on C34 using S‐adenosylmethionine (SAM) as methyl group donor, and this can be further oxidised by ABH1 in the presence of O2, Fe(II) (Fe2+) and alpha‐ketoglutarate (αKG) to produce f5C34.
Comment in
-
Methionine on the rise: how mitochondria changed their codon usage.EMBO J. 2016 Oct 4;35(19):2066-2067. doi: 10.15252/embj.201695385. Epub 2016 Aug 30. EMBO J. 2016. PMID: 27578810 Free PMC article.
References
-
- Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 - PubMed
-
- Agris PF, Vendeix FA, Graham WD (2007) tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 - PubMed
-
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Hölter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus‐Durner V, Hrabĕ de Angelis M, Káradóttir RT et al (2014) Aberrant methylation of tRNAs links cellular stress to neuro‐developmental disorders. EMBO J 33: 2020–2039 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

