In vitro promoter recognition by the catalytic subunit of plant phage-type RNA polymerases
- PMID: 27497992
- PMCID: PMC5040748
- DOI: 10.1007/s11103-016-0518-z
In vitro promoter recognition by the catalytic subunit of plant phage-type RNA polymerases
Abstract
We identified sequence motifs, which enhance or reduce the ability of the Arabidopsis phage-type RNA polymerases RPOTm (mitochondrial RNAP), RPOTp (plastidial RNAP), and RPOTmp (active in both organelles) to recognize their promoters in vitro with help of a 'specificity loop'. The importance of this data for the evolution and function of the organellar RNA polymerases is discussed. The single-subunit RNA polymerase (RNAP) of bacteriophage T7 is able to perform all steps of transcription without additional transcription factors. Dicotyledonous plants possess three phage-type RNAPs, RPOTm-the mitochondrial RNAP, RPOTp-the plastidial RNAP, and RPOTmp-an RNAP active in both organelles. RPOTm and RPOTp, like the T7 polymerase, are able to recognize promoters, while RPOTmp displays no significant promoter specificity in vitro. To find out which promoter motifs are crucial for recognition by the polymerases we performed in vitro transcription assays with recombinant Arabidopsis RPOTm and RPOTp enzymes. By comparing different truncated and mutagenized promoter constructs, we observed the same minimal promoter sequence supposed to be needed in vivo for transcription initiation. Moreover, we identified elements of core and flanking sequences, which are of critical importance for promoter recognition and activity in vitro. We further intended to reveal why RPOTmp does not efficiently recognize promoters in vitro and if promoter recognition is based on a structurally defined specificity loop of the plant enzymes as described for the yeast and T7 RNAPs. Interestingly, the exchange of only three amino acids within the putative specificity loop of RPOTmp enabled the enzyme for specific promoter transcription in vitro. Thus, also in plant phage-type RNAPs the specificity loop is engaged in promoter recognition. The results are discussed with respect to their relevance for transcription in organello and to the evolution of RPOT enzymes including the divergence of their functions.
Keywords: Arabidopsis; Chloroplast RNA polymerase; Mitochondrial RNA polymerase; Phage-type RNA polymerase; Promoter recognition.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






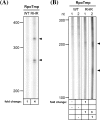
Similar articles
-
Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes.Plant Cell. 2007 Mar;19(3):959-71. doi: 10.1105/tpc.106.046839. Epub 2007 Mar 30. Plant Cell. 2007. PMID: 17400896 Free PMC article.
-
Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases.Planta. 2006 Apr;223(5):998-1009. doi: 10.1007/s00425-005-0159-y. Epub 2005 Nov 24. Planta. 2006. PMID: 16307282
-
Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis.Plant Physiol. 2007 Nov;145(3):712-21. doi: 10.1104/pp.107.103846. Epub 2007 Sep 20. Plant Physiol. 2007. PMID: 17885088 Free PMC article.
-
Chloroplast RNA polymerases: Role in chloroplast biogenesis.Biochim Biophys Acta. 2015 Sep;1847(9):761-9. doi: 10.1016/j.bbabio.2015.02.004. Epub 2015 Feb 11. Biochim Biophys Acta. 2015. PMID: 25680513 Review.
-
Mitochondrial transcription initiation: promoter structures and RNA polymerases.Curr Genet. 1995 Aug;28(3):205-16. doi: 10.1007/BF00309779. Curr Genet. 1995. PMID: 8529266 Review.
Cited by
-
Predicting Evolution of the Transcription Regulatory Network in a Bacteriophage.Genome Biol Evol. 2018 Oct 1;10(10):2614-2628. doi: 10.1093/gbe/evy191. Genome Biol Evol. 2018. PMID: 30184065 Free PMC article.
-
Structure-Function Analysis Reveals the Singularity of Plant Mitochondrial DNA Replication Components: A Mosaic and Redundant System.Plants (Basel). 2019 Nov 21;8(12):533. doi: 10.3390/plants8120533. Plants (Basel). 2019. PMID: 31766564 Free PMC article. Review.
References
-
- Backert S, Nielsen BL, Börner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997;2:477–483. doi: 10.1016/S1360-1385(97)01148-5. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

