Eukaryotic resectosomes: A single-molecule perspective
- PMID: 27498169
- PMCID: PMC5290259
- DOI: 10.1016/j.pbiomolbio.2016.08.001
Eukaryotic resectosomes: A single-molecule perspective
Abstract
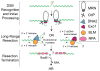
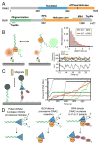
DNA double-strand breaks (DSBs) disrupt the physical and genetic continuity of the genome. If unrepaired, DSBs can lead to cellular dysfunction and malignant transformation. Homologous recombination (HR) is a universally conserved DSB repair mechanism that employs the information in a sister chromatid to catalyze error-free DSB repair. To initiate HR, cells assemble the resectosome: a multi-protein complex composed of helicases, nucleases, and regulatory proteins. The resectosome nucleolytically degrades (resects) the free DNA ends for downstream homologous recombination. Several decades of intense research have identified the core resectosome components in eukaryotes, archaea, and bacteria. More recently, these proteins have been characterized via single-molecule approaches. Here, we focus on recent single-molecule studies that have begun to unravel how nucleases, helicases, processivity factors, and other regulatory proteins dictate the extent and efficiency of DNA resection in eukaryotic cells. We conclude with a discussion of outstanding questions that can be addressed via single-molecule approaches.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

