Regulation of proinflammatory genes by the circulating microRNA hsa-miR-939
- PMID: 27498764
- PMCID: PMC4976376
- DOI: 10.1038/srep30976
Regulation of proinflammatory genes by the circulating microRNA hsa-miR-939
Abstract
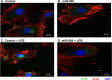
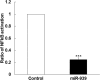
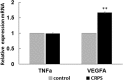
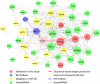
Circulating microRNAs are beneficial biomarkers because of their stability and dysregulation in diseases. Here we sought to determine the role of miR-939, a miRNA downregulated in patients with complex regional pain syndrome (CRPS). Hsa-miR-939 is predicted to target several proinflammatory genes, including IL-6, VEGFA, TNFα, NFκB2, and nitric oxide synthase 2 (NOS2A). Binding of miR-939 to the 3' untranslated region of these genes was confirmed by reporter assay. Overexpression of miR-939 in vitro resulted in reduction of IL-6, NOS2A and NFκB2 mRNAs, IL-6, VEGFA, and NOS2 proteins and NFκB activation. We observed a significant decrease in the NOS substrate l-arginine in plasma from CRPS patients, suggesting reduced miR-939 levels may contribute to an increase in endogenous NOS2A levels and NO, and thereby to pain and inflammation. Pathway analysis showed that miR-939 represents a critical regulatory node in a network of inflammatory mediators. Collectively, our data suggest that miR-939 may regulate multiple proinflammatory genes and that downregulation of miR-939 in CRPS patients may increase expression of these genes, resulting in amplification of the inflammatory pain signal transduction cascade. Circulating miRNAs may function as crucial signaling nodes, and small changes in miRNA levels may influence target gene expression and thus disease.
Figures


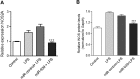

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

