Genomic characterization of the Atlantic cod sex-locus
- PMID: 27499266
- PMCID: PMC4976360
- DOI: 10.1038/srep31235
Genomic characterization of the Atlantic cod sex-locus
Abstract
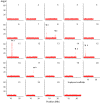
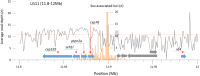
A variety of sex determination mechanisms can be observed in evolutionary divergent teleosts. Sex determination is genetic in Atlantic cod (Gadus morhua), however the genomic location or size of its sex-locus is unknown. Here, we characterize the sex-locus of Atlantic cod using whole genome sequence (WGS) data of 227 wild-caught specimens. Analyzing more than 55 million polymorphic loci, we identify 166 loci that are associated with sex. These loci are located in six distinct regions on five different linkage groups (LG) in the genome. The largest of these regions, an approximately 55 Kb region on LG11, contains the majority of genotypes that segregate closely according to a XX-XY system. Genotypes in this region can be used genetically determine sex, whereas those in the other regions are inconsistently sex-linked. The identified region on LG11 and its surrounding genes have no clear sequence homology with genes or regulatory elements associated with sex-determination or differentiation in other species. The functionality of this sex-locus therefore remains unknown. The WGS strategy used here proved adequate for detecting the small regions associated with sex in this species. Our results highlight the evolutionary flexibility in genomic architecture underlying teleost sex-determination and allow practical applications to genetically sex Atlantic cod.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

