Defective proviruses rapidly accumulate during acute HIV-1 infection
- PMID: 27500724
- PMCID: PMC5014606
- DOI: 10.1038/nm.4156
Defective proviruses rapidly accumulate during acute HIV-1 infection
Abstract
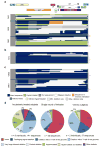
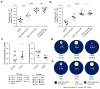
Although antiretroviral therapy (ART) suppresses viral replication to clinically undetectable levels, human immunodeficiency virus type 1 (HIV-1) persists in CD4(+) T cells in a latent form that is not targeted by the immune system or by ART. This latent reservoir is a major barrier to curing individuals of HIV-1 infection. Many individuals initiate ART during chronic infection, and in this setting, most proviruses are defective. However, the dynamics of the accumulation and the persistence of defective proviruses during acute HIV-1 infection are largely unknown. Here we show that defective proviruses accumulate rapidly within the first few weeks of infection to make up over 93% of all proviruses, regardless of how early ART is initiated. By using an unbiased method to amplify near-full-length proviral genomes from HIV-1-infected adults treated at different stages of infection, we demonstrate that early initiation of ART limits the size of the reservoir but does not profoundly affect the proviral landscape. This analysis allows us to revise our understanding of the composition of proviral populations and estimate the true reservoir size in individuals who were treated early versus late in infection. Additionally, we demonstrate that common assays for measuring the reservoir do not correlate with reservoir size, as determined by the number of genetically intact proviruses. These findings reveal hurdles that must be overcome to successfully analyze future HIV-1 cure strategies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. - PubMed
-
- Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. - PubMed
-
- Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

