The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent
- PMID: 27500737
- PMCID: PMC4976988
- DOI: 10.1371/journal.ppat.1005809
The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent
Abstract
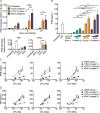
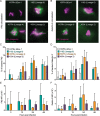
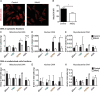
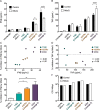
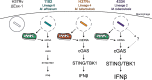
Type I interferons (including IFNαβ) are innate cytokines that may contribute to pathogenesis during Mycobacterium tuberculosis (Mtb) infection. To induce IFNβ, Mtb must gain access to the host cytosol and trigger stimulator of interferon genes (STING) signaling. A recently proposed model suggests that Mtb triggers STING signaling through bacterial DNA binding cyclic GMP-AMP synthase (cGAS) in the cytosol. The aim of this study was to test the generalizability of this model using phylogenetically distinct strains of the Mtb complex (MTBC). We infected bone marrow derived macrophages with strains from MTBC Lineages 2, 4 and 6. We found that the Lineage 6 strain induced less IFNβ, and that the Lineage 2 strain induced more IFNβ, than the Lineage 4 strain. The strains did not differ in their access to the host cytosol and IFNβ induction by each strain required both STING and cGAS. We also found that the three strains shed similar amounts of bacterial DNA. Interestingly, we found that the Lineage 6 strain was associated with less mitochondrial stress and less mitochondrial DNA (mtDNA) in the cytosol compared with the Lineage 4 strain. Treating macrophages with a mitochondria-specific antioxidant reduced cytosolic mtDNA and inhibited IFNβ induction by the Lineage 2 and 4 strains. We also found that the Lineage 2 strain did not induce more mitochondrial stress than the Lineage 4 strain, suggesting that additional pathways contribute to higher IFNβ induction. These results indicate that the mechanism for IFNβ by Mtb is more complex than the established model suggests. We show that mitochondrial dynamics and mtDNA contribute to IFNβ induction by Mtb. Moreover, we show that the contribution of mtDNA to the IFNβ response varies by MTBC strain and that additional mechanisms exist for Mtb to induce IFNβ.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

