Detection of Serum microRNAs From Department of Defense Serum Repository: Correlation With Cotinine, Cytokine, and Polycyclic Aromatic Hydrocarbon Levels
- PMID: 27501106
- PMCID: PMC5661968
- DOI: 10.1097/JOM.0000000000000742
Detection of Serum microRNAs From Department of Defense Serum Repository: Correlation With Cotinine, Cytokine, and Polycyclic Aromatic Hydrocarbon Levels
Abstract
Objective: The aim of this study was to investigate whether serum samples from the Department of Defense Serum Repository (DoDSR) are of sufficient quality to detect microRNAs (miRNAs), cytokines, immunoglobulin E (IgE), and polycyclic aromatic hydrocarbons (PAHs).
Methods: MiRNAs were isolated and quantified by polymerase chain reaction (PCR) array. Cytokines and chemokines related to inflammation were measured using multiplex immunoassays. Cotinine and IgE were detected by enzyme-linked immunoassay (ELISA) and PAHs were detected by Liquid Chromatography/Mass Spectroscopy.
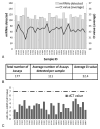
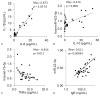
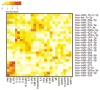
Results: We detected miRNAs, cytokines, IgE, and PAHs with high sensitivity. Eleven of 30 samples tested positive for cotinine suggesting tobacco exposure. Significant associations between serum cotinine, cytokine, IgE, PAHs, and miRNA were discovered.
Conclusion: We successfully quantified over 200 potential biomarkers of occupational exposure from DoDSR samples. The stored serum samples were not affected by hemolysis and represent a powerful tool for biomarker discovery and analysis in retrospective studies.
Conflict of interest statement
There are no conflicts of interest.
Figures



Similar articles
-
MicroRNA Expression Profiling of the Armed Forces Health Surveillance Branch Cohort for Identification of "Enviro-miRs" Associated With Deployment-Based Environmental Exposure.J Occup Environ Med. 2016 Aug;58(8 Suppl 1):S97-S103. doi: 10.1097/JOM.0000000000000764. J Occup Environ Med. 2016. PMID: 27501110
-
Polycyclic aromatic hydrocarbons (PAHs) as determinants of various anthropometric measures of birth outcome.Sci Total Environ. 2013 Feb 1;444:565-78. doi: 10.1016/j.scitotenv.2012.12.021. Epub 2013 Jan 9. Sci Total Environ. 2013. PMID: 23314068
-
Predicting risk of low birth weight offspring from maternal features and blood polycyclic aromatic hydrocarbon concentration.Reprod Toxicol. 2020 Jun;94:92-100. doi: 10.1016/j.reprotox.2020.03.009. Epub 2020 Apr 10. Reprod Toxicol. 2020. PMID: 32283251
-
Urinary carcinogenic 4-6 ring polycyclic aromatic hydrocarbons in coke oven workers and in subjects belonging to the general population: role of occupational and environmental exposure.Int J Hyg Environ Health. 2014 Mar;217(2-3):231-8. doi: 10.1016/j.ijheh.2013.06.005. Epub 2013 Jun 20. Int J Hyg Environ Health. 2014. PMID: 23867119
-
MicroRNAs as Novel Biomarkers of Deployment Status and Exposure to Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans.J Occup Environ Med. 2016 Aug;58(8 Suppl 1):S89-96. doi: 10.1097/JOM.0000000000000769. J Occup Environ Med. 2016. PMID: 27501109 Free PMC article.
Cited by
-
Machine Learning Approach for Predicting Past Environmental Exposures From Molecular Profiling of Post-Exposure Human Serum Samples.J Occup Environ Med. 2019 Dec;61 Suppl 12(Suppl 12):S55-S64. doi: 10.1097/JOM.0000000000001692. J Occup Environ Med. 2019. PMID: 31800451 Free PMC article.
-
Associations of Benzo(ghi)perylene and Heptachlorodibenzo-p-dioxin in Serum of Service Personnel Deployed to Balad, Iraq, and Bagram, Afghanistan Correlates With Perturbed Amino Acid Metabolism in Human Lung Fibroblasts.J Occup Environ Med. 2019 Dec;61 Suppl 12(Suppl 12):S35-S44. doi: 10.1097/JOM.0000000000001669. J Occup Environ Med. 2019. PMID: 31800449 Free PMC article.
-
Polycyclic Aromatic Hydrocarbons and Polychlorinated Dibenzo-p-Dioxins/Dibenzofurans in Microliter Samples of Human Serum as Exposure Indicators.J Occup Environ Med. 2016 Aug;58(8 Suppl 1):S72-9. doi: 10.1097/JOM.0000000000000743. J Occup Environ Med. 2016. PMID: 27501107 Free PMC article.
-
Exposure to Heptachlorodibenzo-p-dioxin (HpCDD) Regulates microRNA Expression in Human Lung Fibroblasts.J Occup Environ Med. 2019 Dec;61 Suppl 12(Suppl 12):S82-S89. doi: 10.1097/JOM.0000000000001691. J Occup Environ Med. 2019. PMID: 31800454 Free PMC article.
-
Implementation of multiomic mass spectrometry approaches for the evaluation of human health following environmental exposure.Mol Omics. 2024 Jun 10;20(5):296-321. doi: 10.1039/d3mo00214d. Mol Omics. 2024. PMID: 38623720 Free PMC article. Review.
References
-
- Smith B, Wong CA, Boyko EJ, Phillips CJ, Gackstetter GD, Ryan MA, et al. The effects of exposure to documented open-air burn pits on respiratory health among deployers of the Millennium Cohort Study. J Occup Environ Med. 2012;54:708–716. - PubMed
-
- Mancuso JD, Mallon TM, Gaydos JC. Maximizing the capabilities of the DoD serum repository to meet current and future needs, report of the needs panel. Mil Med. 2015;180(10 Suppl):13–24. - PubMed
-
- Grasedieck S, Scholer N, Bommer M, Niess JH, Tumani H, Rouhi A, et al. Impact of serum storage conditions on microRNA stability. Leukemia. 2012;26:2414–2416. - PubMed
-
- Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

