Differential Assembly of Catalytic Interactions within the Conserved Active Sites of Two Ribozymes
- PMID: 27501145
- PMCID: PMC4976970
- DOI: 10.1371/journal.pone.0160457
Differential Assembly of Catalytic Interactions within the Conserved Active Sites of Two Ribozymes
Abstract
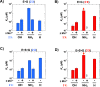
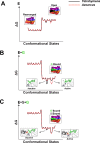
Molecular recognition is central to biology and a critical aspect of RNA function. Yet structured RNAs typically lack the preorganization needed for strong binding and precise positioning. A striking example is the group I ribozyme from Tetrahymena, which binds its guanosine substrate (G) orders of magnitude slower than diffusion. Binding of G is also thermodynamically coupled to binding of the oligonucleotide substrate (S) and further work has shown that the transition from E•G to E•S•G accompanies a conformational change that allows G to make the active site interactions required for catalysis. The group I ribozyme from Azoarcus has a similarly slow association rate but lacks the coupled binding observed for the Tetrahymena ribozyme. Here we test, using G analogs and metal ion rescue experiments, whether this absence of coupling arises from a higher degree of preorganization within the Azoarcus active site. Our results suggest that the Azoarcus ribozyme forms cognate catalytic metal ion interactions with G in the E•G complex, interactions that are absent in the Tetrahymena E•G complex. Thus, RNAs that share highly similar active site architectures and catalyze the same reactions can differ in the assembly of transition state interactions. More generally, an ability to readily access distinct local conformational states may have facilitated the evolutionary exploration needed to attain RNA machines that carry out complex, multi-step processes.
Conflict of interest statement
Figures

Similar articles
-
Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2'-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme.Biochemistry. 1999 Aug 24;38(34):10958-75. doi: 10.1021/bi990388e. Biochemistry. 1999. PMID: 10460151
-
A kinetic and thermodynamic framework for the Azoarcus group I ribozyme reaction.RNA. 2014 Nov;20(11):1732-46. doi: 10.1261/rna.044362.114. Epub 2014 Sep 22. RNA. 2014. PMID: 25246656 Free PMC article.
-
Leaving group stabilization by metal ion coordination and hydrogen bond donation is an evolutionarily conserved feature of group I introns.Biochim Biophys Acta. 2001 Dec 30;1522(3):158-66. doi: 10.1016/s0167-4781(01)00327-x. Biochim Biophys Acta. 2001. PMID: 11779630
-
Identification of catalytic metal ion ligands in ribozymes.Methods. 2009 Oct;49(2):148-66. doi: 10.1016/j.ymeth.2009.07.005. Epub 2009 Aug 3. Methods. 2009. PMID: 19651216 Free PMC article. Review.
-
The structure and function of catalytic RNAs.Sci China C Life Sci. 2009 Mar;52(3):232-44. doi: 10.1007/s11427-009-0038-z. Epub 2009 Mar 18. Sci China C Life Sci. 2009. PMID: 19294348 Review.
References
-
- Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7: 834–7. - PubMed
-
- Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287: 820–5. - PubMed
-
- Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6: 399–412. - PubMed
-
- Sigler PB. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4: 477–527. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

