FRET binding antenna reports spatiotemporal dynamics of GDI-Cdc42 GTPase interactions
- PMID: 27501396
- PMCID: PMC5030135
- DOI: 10.1038/nchembio.2145
FRET binding antenna reports spatiotemporal dynamics of GDI-Cdc42 GTPase interactions
Abstract
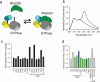
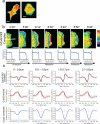
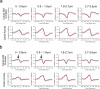
Guanine-nucleotide dissociation inhibitors (GDIs) are negative regulators of Rho family GTPases that sequester the GTPases away from the membrane. Here we ask how GDI-Cdc42 interaction regulates localized Cdc42 activation for cell motility. The sensitivity of cells to overexpression of Rho family pathway components led us to a new biosensor, GDI.Cdc42 FLARE, in which Cdc42 is modified with a fluorescence resonance energy transfer (FRET) 'binding antenna' that selectively reports Cdc42 binding to endogenous GDIs. Similar antennae could also report GDI-Rac1 and GDI-RhoA interaction. Through computational multiplexing and simultaneous imaging, we determined the spatiotemporal dynamics of GDI-Cdc42 interaction and Cdc42 activation during cell protrusion and retraction. This revealed remarkably tight coordination of GTPase release and activation on a time scale of 10 s, suggesting that GDI-Cdc42 interactions are a critical component of the spatiotemporal regulation of Cdc42 activity, and not merely a mechanism for global sequestration of an inactivated pool of signaling molecules.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

