AraC-Type Regulator Rbf Controls the Staphylococcus epidermidis Biofilm Phenotype by Negatively Regulating the icaADBC Repressor SarR
- PMID: 27501984
- PMCID: PMC5055596
- DOI: 10.1128/JB.00374-16
AraC-Type Regulator Rbf Controls the Staphylococcus epidermidis Biofilm Phenotype by Negatively Regulating the icaADBC Repressor SarR
Abstract
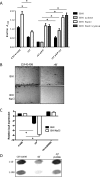
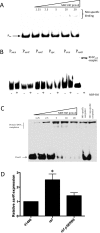
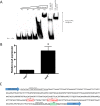
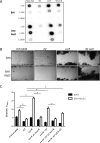
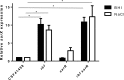
Regulation of icaADBC-encoded polysaccharide intercellular adhesin (PIA)/poly-N-acetylglucosasmine (PNAG) production in staphylococci plays an important role in biofilm-associated medical-device-related infections. Here, we report that the AraC-type transcriptional regulator Rbf activates icaADBC operon transcription and PIA production in Staphylococcus epidermidis Purified recombinant Rbf did not bind to the ica operon promoter region in electrophoretic mobility shift assays (EMSAs), indicating that Rbf regulates ica transcription indirectly. To identify the putative transcription factor(s) involved in Rbf-mediated icaADBC regulation, the ability of recombinant Rbf to interact with the promoter sequences of known icaADBC regulators was investigated. Recombinant Rbf bound to the sarR promoter and not the sarX, sarA, sarZ, spx, and srrA promoters. Reverse transcription (RT)-PCR demonstrated that Rbf acts as a repressor of sarR transcription. PIA expression and biofilm production were restored to wild-type levels in an rbf sarR double mutant grown in brain heart infusion (BHI) medium supplemented with NaCl, which is known to activate the ica locus, but not in BHI medium alone. RT-PCR further demonstrated that although Rbf does not bind the sarX promoter, it nevertheless exerted a negative effect on sarX expression. Apparently, direct downregulation of the SarR repressor by Rbf has a dominant effect over indirect repression of the SarX activator by Rbf in the control of S. epidermidis PIA production and biofilm formation.
Importance: The importance of Staphylococcus epidermidis as an opportunistic pathogen in hospital patients with implanted medical devices derives largely from its capacity to form biofilm. Expression of the icaADBC-encoded extracellular polysaccharide is the predominant biofilm mechanism in S. epidermidis clinical isolates and is tightly regulated. Here, we report that the transcriptional regulator Rbf promotes icaADBC expression by negatively regulating expression of sarR, which encodes an ica operon repressor. Furthermore, Rbf indirectly represses the ica operon activator, SarX. The data reveal complicated interplay between Rbf and two Sar family proteins in fine-tuning regulation of the biofilm phenotype and indicate that in the hierarchy of biofilm regulators, IcaR is dominant over the Rbf-SarR-SarX axis.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
A novel role for SarX in Staphylococcus epidermidis biofilm regulation.Microbiology (Reading). 2011 Apr;157(Pt 4):1042-1049. doi: 10.1099/mic.0.046581-0. Epub 2011 Feb 3. Microbiology (Reading). 2011. PMID: 21292751
-
SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development.J Bacteriol. 2005 Apr;187(7):2348-56. doi: 10.1128/JB.187.7.2348-2356.2005. J Bacteriol. 2005. PMID: 15774878 Free PMC article.
-
SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis.Can J Microbiol. 2007 Jan;53(1):82-91. doi: 10.1139/w06-108. Can J Microbiol. 2007. PMID: 17496953
-
Genetic regulation of the intercellular adhesion locus in staphylococci.Front Cell Infect Microbiol. 2012 Mar 26;2:38. doi: 10.3389/fcimb.2012.00038. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 23061050 Free PMC article. Review.
-
Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects.Front Cell Infect Microbiol. 2015 Feb 10;5:7. doi: 10.3389/fcimb.2015.00007. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25713785 Free PMC article. Review.
Cited by
-
Small RNA teg49 Is Derived from a sarA Transcript and Regulates Virulence Genes Independent of SarA in Staphylococcus aureus.Infect Immun. 2018 Jan 22;86(2):e00635-17. doi: 10.1128/IAI.00635-17. Print 2018 Feb. Infect Immun. 2018. PMID: 29133345 Free PMC article.
-
Complete Genome Sequence of Staphylococcus epidermidis 1457.Genome Announc. 2017 Jun 1;5(22):e00450-17. doi: 10.1128/genomeA.00450-17. Genome Announc. 2017. PMID: 28572323 Free PMC article.
-
TMT proteomic analysis for molecular mechanism of Staphylococcus aureus in response to freezing stress.Appl Microbiol Biotechnol. 2022 Apr;106(8):3139-3152. doi: 10.1007/s00253-022-11927-w. Epub 2022 Apr 23. Appl Microbiol Biotechnol. 2022. PMID: 35460349
-
BfvR, an AraC-Family Regulator, Controls Biofilm Formation and pH6 Antigen Production in Opposite Ways in Yersinia pestis Biovar Microtus.Front Cell Infect Microbiol. 2018 Oct 2;8:347. doi: 10.3389/fcimb.2018.00347. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30333962 Free PMC article.
-
Genomic comparisons and phylogenetic analysis of mastitis-related staphylococci with a focus on adhesion, biofilm, and related regulatory genes.Sci Rep. 2021 Aug 30;11(1):17392. doi: 10.1038/s41598-021-96842-2. Sci Rep. 2021. PMID: 34462461 Free PMC article.
References
-
- Hogan S, Stevens NT, Humphreys H, O'Gara JP, O'Neill E. 2015. Current and future approaches to the prevention and treatment of staphylococcal medical device-related infections. Curr Pharm Des 21:100–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

