Radiogenomics to characterize regional genetic heterogeneity in glioblastoma
- PMID: 27502248
- PMCID: PMC5193022
- DOI: 10.1093/neuonc/now135
Radiogenomics to characterize regional genetic heterogeneity in glioblastoma
Abstract
Background: Glioblastoma (GBM) exhibits profound intratumoral genetic heterogeneity. Each tumor comprises multiple genetically distinct clonal populations with different therapeutic sensitivities. This has implications for targeted therapy and genetically informed paradigms. Contrast-enhanced (CE)-MRI and conventional sampling techniques have failed to resolve this heterogeneity, particularly for nonenhancing tumor populations. This study explores the feasibility of using multiparametric MRI and texture analysis to characterize regional genetic heterogeneity throughout MRI-enhancing and nonenhancing tumor segments.
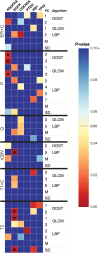
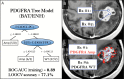
Methods: We collected multiple image-guided biopsies from primary GBM patients throughout regions of enhancement (ENH) and nonenhancing parenchyma (so called brain-around-tumor, [BAT]). For each biopsy, we analyzed DNA copy number variants for core GBM driver genes reported by The Cancer Genome Atlas. We co-registered biopsy locations with MRI and texture maps to correlate regional genetic status with spatially matched imaging measurements. We also built multivariate predictive decision-tree models for each GBM driver gene and validated accuracies using leave-one-out-cross-validation (LOOCV).
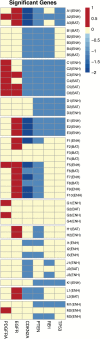
Results: We collected 48 biopsies (13 tumors) and identified significant imaging correlations (univariate analysis) for 6 driver genes: EGFR, PDGFRA, PTEN, CDKN2A, RB1, and TP53. Predictive model accuracies (on LOOCV) varied by driver gene of interest. Highest accuracies were observed for PDGFRA (77.1%), EGFR (75%), CDKN2A (87.5%), and RB1 (87.5%), while lowest accuracy was observed in TP53 (37.5%). Models for 4 driver genes (EGFR, RB1, CDKN2A, and PTEN) showed higher accuracy in BAT samples (n = 16) compared with those from ENH segments (n = 32).
Conclusion: MRI and texture analysis can help characterize regional genetic heterogeneity, which offers potential diagnostic value under the paradigm of individualized oncology.
Keywords: genetic; glioblastoma; heterogeneity; radiogenomics; texture.
© The Author(s) 2016. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

