Canalization of gene expression is a major signature of regulatory cold adaptation in temperate Drosophila melanogaster
- PMID: 27502401
- PMCID: PMC4977637
- DOI: 10.1186/s12864-016-2866-0
Canalization of gene expression is a major signature of regulatory cold adaptation in temperate Drosophila melanogaster
Abstract
Background: Transcriptome analysis may provide means to investigate the underlying genetic causes of shared and divergent phenotypes in different populations and help to identify potential targets of adaptive evolution. Applying RNA sequencing to whole male Drosophila melanogaster from the ancestral tropical African environment and a very recently colonized cold-temperate European environment at both standard laboratory conditions and following a cold shock, we seek to uncover the transcriptional basis of cold adaptation.
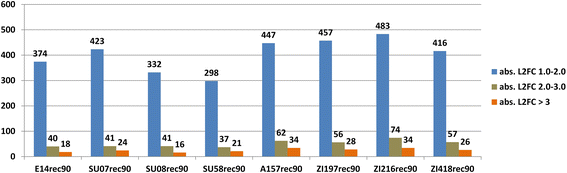
Results: In both the ancestral and the derived populations, the predominant characteristic of the cold shock response is the swift and massive upregulation of heat shock proteins and other chaperones. Although we find ~25 % of the genome to be differentially expressed following a cold shock, only relatively few genes (n = 16) are up- or down-regulated in a population-specific way. Intriguingly, 14 of these 16 genes show a greater degree of differential expression in the African population. Likewise, there is an excess of genes with particularly strong cold-induced changes in expression in Africa on a genome-wide scale.
Conclusions: The analysis of the transcriptional cold shock response most prominently reveals an upregulation of components of a general stress response, which is conserved over many taxa and triggered by a plethora of stressors. Despite the overall response being fairly similar in both populations, there is a definite excess of genes with a strong cold-induced fold-change in Africa. This is consistent with a detrimental deregulation or an overshooting stress response. Thus, the canalization of European gene expression might be responsible for the increased cold tolerance of European flies.
Keywords: Adaptation; Canalization; Cold tolerance; Transcriptomics.
Figures



Similar articles
-
De novo transcriptome sequencing and gene expression profiling of Elymus nutans under cold stress.BMC Genomics. 2016 Nov 4;17(1):870. doi: 10.1186/s12864-016-3222-0. BMC Genomics. 2016. PMID: 27814694 Free PMC article.
-
Transcriptome Profiling of Two Asparagus Bean (Vigna unguiculata subsp. sesquipedalis) Cultivars Differing in Chilling Tolerance under Cold Stress.PLoS One. 2016 Mar 8;11(3):e0151105. doi: 10.1371/journal.pone.0151105. eCollection 2016. PLoS One. 2016. PMID: 26954786 Free PMC article.
-
The Role of Inducible Hsp70, and Other Heat Shock Proteins, in Adaptive Complex of Cold Tolerance of the Fruit Fly (Drosophila melanogaster).PLoS One. 2015 Jun 2;10(6):e0128976. doi: 10.1371/journal.pone.0128976. eCollection 2015. PLoS One. 2015. PMID: 26034990 Free PMC article.
-
Population and sex differences in Drosophila melanogaster brain gene expression.BMC Genomics. 2012 Nov 21;13:654. doi: 10.1186/1471-2164-13-654. BMC Genomics. 2012. PMID: 23170910 Free PMC article.
-
Systemic analysis of stress transcriptomics of Synechocystis reveals common stress genes and their universal triggers.Mol Biosyst. 2016 Oct 18;12(11):3254-3258. doi: 10.1039/c6mb00551a. Mol Biosyst. 2016. PMID: 27754509 Review.
Cited by
-
Metabolically similar cohorts of bacteria exhibit strong cooccurrence patterns with diet items and eukaryotic microbes in lizard guts.Ecol Evol. 2019 Oct 23;9(22):12471-12481. doi: 10.1002/ece3.5691. eCollection 2019 Nov. Ecol Evol. 2019. PMID: 31788191 Free PMC article.
-
Gene Regulatory Evolution in Cold-Adapted Fly Populations Neutralizes Plasticity and May Undermine Genetic Canalization.Genome Biol Evol. 2022 Apr 10;14(4):evac050. doi: 10.1093/gbe/evac050. Genome Biol Evol. 2022. PMID: 35380655 Free PMC article.
-
Parallel and population-specific gene regulatory evolution in cold-adapted fly populations.Genetics. 2021 Jul 14;218(3):iyab077. doi: 10.1093/genetics/iyab077. Genetics. 2021. PMID: 33989401 Free PMC article.
-
Three Quantitative Trait Loci Explain More than 60% of Variation for Chill Coma Recovery Time in a Natural Population of Drosophila ananassae.G3 (Bethesda). 2019 Nov 5;9(11):3715-3725. doi: 10.1534/g3.119.400453. G3 (Bethesda). 2019. PMID: 31690597 Free PMC article.
-
Genomic Shifts, Phenotypic Clines, and Fitness Costs Associated With Cold Tolerance in the Asian Tiger Mosquito.Mol Biol Evol. 2022 May 3;39(5):msac104. doi: 10.1093/molbev/msac104. Mol Biol Evol. 2022. PMID: 35574643 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

