Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation
- PMID: 27503198
- PMCID: PMC5467311
- DOI: 10.1158/1078-0432.CCR-16-0594
Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation
Abstract
Purpose: Chronic lymphocytic leukemia (CLL) with 17p deletion typically progresses quickly and is refractory to most conventional therapies. However, some del(17p) patients do not progress for years, suggesting that del(17p) is not the only driving event in CLL progression. We hypothesize that other concomitant genetic abnormalities underlie the clinical heterogeneity of del(17p) CLL.
Experimental design: We profiled the somatic mutations and copy number alterations (CNA) in a large group of del(17p) CLLs as well as wild-type CLL and analyzed the genetic basis of their clinical heterogeneity.
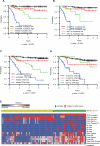
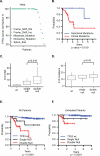
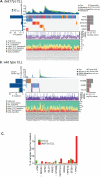
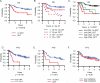
Results: We found that increased somatic mutation number associates with poor overall survival independent of 17p deletion (P = 0.003). TP53 mutation was present in 81% of del(17p) CLL, mostly clonal (82%), and clonal mutations with del(17p) exhibit shorter overall survival than subclonal mutations with del(17p) (P = 0.019). Del(17p) CLL has a unique driver mutation profile, including NOTCH1 (15%), RPS15 (12%), DDX3X (8%), and GPS2 (6%). We found that about half of del(17p) CLL cases have recurrent deletions at 3p, 4p, or 9p and that any of these deletions significantly predicts shorter overall survival. In addition, the number of CNAs, but not somatic mutations, predicts shorter time to treatment among patients untreated at sampling. Indolent del(17p) CLLs were characterized by absent or subclonal TP53 mutation and few CNAs, with no difference in somatic mutation number.
Conclusions: We conclude that del(17p) has a unique genomic profile and that clonal TP53 mutations, 3p, 4p, or 9p deletions, and genomic complexity are associated with shorter overall survival. Clin Cancer Res; 23(3); 735-45. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures
References
-
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. - PubMed
-
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. - PubMed
-
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. - PubMed
-
- Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21:2442–51. - PubMed
-
- Baliakas P, Iskas M, Gardiner A, Davis Z, Plevova K, Nguyen-Khac F, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89:249–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

