Comparability of automated human induced pluripotent stem cell culture: a pilot study
- PMID: 27503483
- PMCID: PMC5050253
- DOI: 10.1007/s00449-016-1659-9
Comparability of automated human induced pluripotent stem cell culture: a pilot study
Abstract
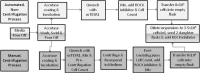
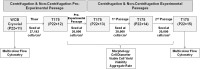
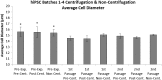
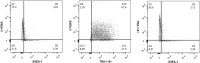
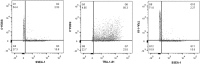
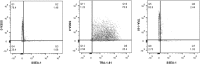
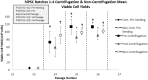
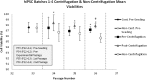
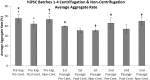
Consistent and robust manufacturing is essential for the translation of cell therapies, and the utilisation automation throughout the manufacturing process may allow for improvements in quality control, scalability, reproducibility and economics of the process. The aim of this study was to measure and establish the comparability between alternative process steps for the culture of hiPSCs. Consequently, the effects of manual centrifugation and automated non-centrifugation process steps, performed using TAP Biosystems' CompacT SelecT automated cell culture platform, upon the culture of a human induced pluripotent stem cell (hiPSC) line (VAX001024c07) were compared. This study, has demonstrated that comparable morphologies and cell diameters were observed in hiPSCs cultured using either manual or automated process steps. However, non-centrifugation hiPSC populations exhibited greater cell yields, greater aggregate rates, increased pluripotency marker expression, and decreased differentiation marker expression compared to centrifugation hiPSCs. A trend for decreased variability in cell yield was also observed after the utilisation of the automated process step. This study also highlights the detrimental effect of the cryopreservation and thawing processes upon the growth and characteristics of hiPSC cultures, and demonstrates that automated hiPSC manufacturing protocols can be successfully transferred between independent laboratories.
Keywords: Automation; Centrifugation; Characterisation; Comparability; Pluripotent stem cell; Scalable.
Conflict of interest statement
This research, in which TAP Biosystems’ CompacT SelecT automated cell culture platform was utilised, was funded by TAP Biosystems as well as Loughborough University and the Engineering and Physical Sciences Research Council (EPSRC). Neither Loughborough University, as a funding body, nor the EPSRC were involved in the preparation of this publication. However, Dave Thomas, Product Manager at TAP Biosystems, advised on study design and data interpretation, in addition to providing research supervision for this work by acting as an industrial supervisor. The terms of this arrangement have been reviewed and approved by Loughborough University.
Figures





References
-
- Archibald PRT, Chandra A, Thomas D, Morley G, Lekishvili T, Devonshire A, Williams DJ. Comparability of scalable, automated hMSC culture using manual and automated process steps. Biochem Eng J. 2016;108:69–83. doi: 10.1016/j.bej.2015.07.001. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

