Regulatory evolution of Tbx5 and the origin of paired appendages
- PMID: 27503876
- PMCID: PMC5018757
- DOI: 10.1073/pnas.1609997113
Regulatory evolution of Tbx5 and the origin of paired appendages
Abstract
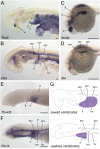
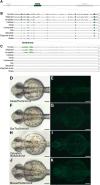
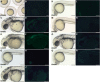
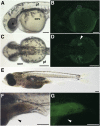
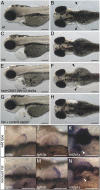
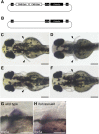
The diversification of paired appendages has been a major factor in the evolutionary radiation of vertebrates. Despite its importance, an understanding of the origin of paired appendages has remained elusive. To address this problem, we focused on T-box transcription factor 5 (Tbx5), a gene indispensable for pectoral appendage initiation and development. Comparison of gene expression in jawless and jawed vertebrates reveals that the Tbx5 expression in jawed vertebrates is derived in having an expression domain that extends caudal to the heart and gills. Chromatin profiling, phylogenetic footprinting, and functional assays enabled the identification of a Tbx5 fin enhancer associated with this apomorphic pattern of expression. Comparative functional analysis of reporter constructs reveals that this enhancer activity is evolutionarily conserved among jawed vertebrates and is able to rescue the finless phenotype of tbx5a mutant zebrafish. Taking paleontological evidence of early vertebrates into account, our results suggest that the gain of apomorphic patterns of Tbx5 expression and regulation likely contributed to the morphological transition from a finless to finned condition at the base of the vertebrate lineage.
Keywords: Tbx5 enhancer; development; evolution; paired fins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Thacher JK. Median and paired fins, a contribution to the history of the vertebrate limbs. Trans Conn Acad Arts Sci. 1877;3:281–310.
-
- Mivart S. Notes on the fins of elasmobranchs, with considerations on the nature and homologues of vertebrate limbs. Trans Zool Soc Lond. 1879;10:439–484.
-
- Balfour FM. On the dedopment of the skeleton of the paired fins of Elasmobranchii, considered in relation to its bearings on the nature of the limbs of the vertebrata. Proc Zool Soc Lond. 1881;49(3):656–671.
-
- Gegenbaur C. Elements of Comparative Anatomy. MacMillan and Co.; London: 1878.
-
- Goodrich E. Studies on the Structure and Development of Vertebrates. MacMillan and Co., Limited; London: 1930.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

