ORM Expression Alters Sphingolipid Homeostasis and Differentially Affects Ceramide Synthase Activity
- PMID: 27506241
- PMCID: PMC5047106
- DOI: 10.1104/pp.16.00965
ORM Expression Alters Sphingolipid Homeostasis and Differentially Affects Ceramide Synthase Activity
Abstract
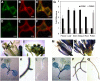
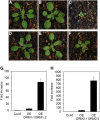
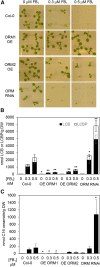
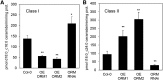
Sphingolipid synthesis is tightly regulated in eukaryotes. This regulation in plants ensures sufficient sphingolipids to support growth while limiting the accumulation of sphingolipid metabolites that induce programmed cell death. Serine palmitoyltransferase (SPT) catalyzes the first step in sphingolipid biosynthesis and is considered the primary sphingolipid homeostatic regulatory point. In this report, Arabidopsis (Arabidopsis thaliana) putative SPT regulatory proteins, orosomucoid-like proteins AtORM1 and AtORM2, were found to interact physically with Arabidopsis SPT and to suppress SPT activity when coexpressed with Arabidopsis SPT subunits long-chain base1 (LCB1) and LCB2 and the small subunit of SPT in a yeast (Saccharomyces cerevisiae) SPT-deficient mutant. Consistent with a role in SPT suppression, AtORM1 and AtORM2 overexpression lines displayed increased resistance to the programmed cell death-inducing mycotoxin fumonisin B1, with an accompanying reduced accumulation of LCBs and C16 fatty acid-containing ceramides relative to wild-type plants. Conversely, RNA interference (RNAi) suppression lines of AtORM1 and AtORM2 displayed increased sensitivity to fumonisin B1 and an accompanying strong increase in LCBs and C16 fatty acid-containing ceramides relative to wild-type plants. Overexpression lines also were found to have reduced activity of the class I ceramide synthase that uses C16 fatty acid acyl-coenzyme A and dihydroxy LCB substrates but increased activity of class II ceramide synthases that use very-long-chain fatty acyl-coenzyme A and trihydroxy LCB substrates. RNAi suppression lines, in contrast, displayed increased class I ceramide synthase activity but reduced class II ceramide synthase activity. These findings indicate that ORM mediation of SPT activity differentially regulates functionally distinct ceramide synthase activities as part of a broader sphingolipid homeostatic regulatory network.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Bach L, Gissot L, Marion J, Tellier F, Moreau P, Satiat-Jeunemaître B, Palauqui JC, Napier JA, Faure JD (2011) Very-long-chain fatty acids are required for cell plate formation during cytokinesis in Arabidopsis thaliana. J Cell Sci 124: 3223–3234 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

