ER fatalities-The role of ER-mitochondrial contact sites in yeast life and death decisions
- PMID: 27507669
- PMCID: PMC5293681
- DOI: 10.1016/j.mad.2016.07.007
ER fatalities-The role of ER-mitochondrial contact sites in yeast life and death decisions
Abstract
Following extracellular stress signals, all eukaryotic cells choose whether to elicit a pro-survival or pro-death response. The decision over which path to take is governed by the severity and duration of the damage. In response to mild stress, pro-survival programs are initiated (unfolded protein response, autophagy, mitophagy) whereas severe or chronic stress forces the cell to abandon these adaptive programs and shift towards regulated cell death to remove irreversibly damaged cells. Both pro-survival and pro-death programs involve regulated communication between the endoplasmic reticulum (ER) and mitochondria. In yeast, recent data suggest this inter-organelle contact is facilitated by the endoplasmic reticulum mitochondria encounter structure (ERMES). These membrane contacts are not only important for the exchange of cellular signals, but also play a role in mitochondrial tethering during mitophagy, mitochondrial fission and mitochondrial inheritance. This review focuses on recent findings in yeast that shed light on how ER-mitochondrial communication mediates critical cell fate decisions.
Keywords: ERMES; Endoplasmic reticulum; Mitochondria; Mitophagy; Programmed cell death; Signal transduction.
Copyright © 2016 Elsevier Ireland Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest
Figures
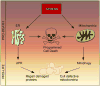


Similar articles
-
Mitophagy in yeast: Molecular mechanisms and physiological role.Biochim Biophys Acta. 2015 Oct;1853(10 Pt B):2756-65. doi: 10.1016/j.bbamcr.2015.01.005. Epub 2015 Jan 17. Biochim Biophys Acta. 2015. PMID: 25603537 Review.
-
Mitochondrial ER contacts are crucial for mitophagy in yeast.Dev Cell. 2014 Feb 24;28(4):450-8. doi: 10.1016/j.devcel.2014.01.012. Epub 2014 Feb 13. Dev Cell. 2014. PMID: 24530295
-
ER-mitochondria contacts as sites of mitophagosome formation.Autophagy. 2014 Jul;10(7):1346-7. doi: 10.4161/auto.28981. Epub 2014 May 15. Autophagy. 2014. PMID: 24905224 Free PMC article.
-
Making connections: interorganelle contacts orchestrate mitochondrial behavior.Trends Cell Biol. 2014 Sep;24(9):537-45. doi: 10.1016/j.tcb.2014.04.004. Epub 2014 Apr 28. Trends Cell Biol. 2014. PMID: 24786308 Review.
-
Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy.Autophagy. 2017 Jan 2;13(1):114-132. doi: 10.1080/15548627.2016.1252889. Epub 2016 Nov 15. Autophagy. 2017. PMID: 27846375 Free PMC article.
Cited by
-
Mechanisms Underlying the Essential Role of Mitochondrial Membrane Lipids in Yeast Chronological Aging.Oxid Med Cell Longev. 2017;2017:2916985. doi: 10.1155/2017/2916985. Epub 2017 May 16. Oxid Med Cell Longev. 2017. PMID: 28593023 Free PMC article. Review.
-
A dynamic actin cytoskeleton is required to prevent constitutive VDAC-dependent MAPK signalling and aberrant lipid homeostasis.iScience. 2023 Aug 2;26(9):107539. doi: 10.1016/j.isci.2023.107539. eCollection 2023 Sep 15. iScience. 2023. PMID: 37636069 Free PMC article.
-
Anhydrobiosis in yeast: role of cortical endoplasmic reticulum protein Ist2 in Saccharomyces cerevisiae cells during dehydration and subsequent rehydration.Antonie Van Leeuwenhoek. 2021 Jul;114(7):1069-1077. doi: 10.1007/s10482-021-01578-8. Epub 2021 Apr 12. Antonie Van Leeuwenhoek. 2021. PMID: 33844120
References
-
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14(1):20–8. - PubMed
-
- Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51(3):794–8. - PubMed
-
- Shutt TE, McBride HM. Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochim Biophys Acta. 2013;1833(2):417–24. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

