Engineering of Helicobacter pylori Dimeric Oxidoreductase DsbK (HP0231)
- PMID: 27507968
- PMCID: PMC4960241
- DOI: 10.3389/fmicb.2016.01158
Engineering of Helicobacter pylori Dimeric Oxidoreductase DsbK (HP0231)
Abstract
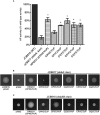
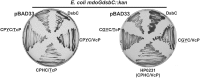
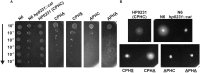
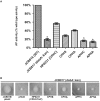
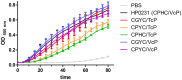
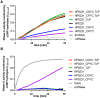
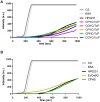
The formation of disulfide bonds that are catalyzed by proteins of the Dsb (disulfide bond) family is crucial for the correct folding of many extracytoplasmic proteins. Thus, this formation plays an essential, pivotal role in the assembly of many virulence factors. The Helicobacter pylori disulfide bond-forming system is uncomplicated compared to the best-characterized Escherichia coli Dsb pathways. It possesses only two extracytoplasmic Dsb proteins named HP0377 and HP0231. As previously shown, HP0377 is a reductase involved in the process of cytochrome c maturation. Additionally, it also possesses disulfide isomerase activity. HP0231 was the first periplasmic dimeric oxidoreductase involved in disulfide generation to be described. Although HP0231 function is critical for oxidative protein folding, its structure resembles that of dimeric EcDsbG, which does not confer this activity. However, the HP0231 catalytic motifs (CXXC and the so-called cis-Pro loop) are identical to that of monomeric EcDsbA. To understand the functioning of HP0231, we decided to study the relations between its sequence, structure and activity through an extensive analysis of various HP0231 point mutants, using in vivo and in vitro strategies. Our work shows the crucial role of the cis-Pro loop, as changing valine to threonine in this motif completely abolishes the protein function in vivo. Functioning of HP0231 is conditioned by the combination of CXXC and the cis-Pro loop, as replacing the HP0231 CXXC motif by the motif from EcDsbG or EcDsbC results in bifunctional protein, at least in E. coli. We also showed that the dimerization domain of HP0231 ensures contact with its substrates. Moreover, the activity of this oxidase is independent on the structure of the catalytic domain. Finally, we showed that HP0231 chaperone activity is independent of its redox function.
Keywords: Dsb proteins; Helicobacter pylori; chaperone activity; disulfide bonds; oxidoreductases; protein engineering; site-directed mutagenesis.
Figures



Similar articles
-
Functional and evolutionary analyses of Helicobacter pylori HP0231 (DsbK) protein with strong oxidative and chaperone activity characterized by a highly diverged dimerization domain.Front Microbiol. 2015 Oct 8;6:1065. doi: 10.3389/fmicb.2015.01065. eCollection 2015. Front Microbiol. 2015. PMID: 26500620 Free PMC article.
-
Helicobacter pylori HP0377, a member of the Dsb family, is an untypical multifunctional CcmG that cooperates with dimeric thioldisulfide oxidase HP0231.BMC Microbiol. 2015 Jul 4;15:135. doi: 10.1186/s12866-015-0471-z. BMC Microbiol. 2015. PMID: 26141380 Free PMC article.
-
Exploring the inhibitory potential of in silico-designed small peptides on Helicobacter pylori Hp0231 (DsbK), a periplasmic oxidoreductase involved in disulfide bond formation.Front Mol Biosci. 2024 Jan 11;10:1335704. doi: 10.3389/fmolb.2023.1335704. eCollection 2023. Front Mol Biosci. 2024. PMID: 38274095 Free PMC article.
-
Engineering of the Dsb (disulfide bond) proteins - contribution towards understanding their mechanism of action and their applications in biotechnology and medicine.Crit Rev Microbiol. 2019 Aug;45(4):433-450. doi: 10.1080/1040841X.2019.1622509. Epub 2019 Jun 13. Crit Rev Microbiol. 2019. PMID: 31190593 Review.
-
Disulfide Bonds: A Key Modification in Bacterial Extracytoplasmic Proteins.J Dent Res. 2017 Dec;96(13):1465-1473. doi: 10.1177/0022034517725059. Epub 2017 Aug 10. J Dent Res. 2017. PMID: 28797211 Review.
Cited by
-
Genomic Features and Tissue Expression Profiles of the Tyrosinase Gene Family in the Chinese Soft-Shelled Turtle (Pelodiscus sinensis).Genes (Basel). 2025 Jul 17;16(7):834. doi: 10.3390/genes16070834. Genes (Basel). 2025. PMID: 40725490 Free PMC article.
-
Bacterial thiol oxidoreductases - from basic research to new antibacterial strategies.Appl Microbiol Biotechnol. 2017 May;101(10):3977-3989. doi: 10.1007/s00253-017-8291-8. Epub 2017 Apr 13. Appl Microbiol Biotechnol. 2017. PMID: 28409380 Free PMC article. Review.
-
Unveiling the versatility of the thioredoxin framework: Insights from the structural examination of Francisella tularensis DsbA1.Comput Struct Biotechnol J. 2024 Nov 22;23:4324-4336. doi: 10.1016/j.csbj.2024.11.034. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39697679 Free PMC article.
-
Impact of selected amino acids of HP0377 (Helicobacter pylori thiol oxidoreductase) on its functioning as a CcmG (cytochrome c maturation) protein and Dsb (disulfide bond) isomerase.PLoS One. 2018 Apr 20;13(4):e0195358. doi: 10.1371/journal.pone.0195358. eCollection 2018. PLoS One. 2018. PMID: 29677198 Free PMC article.
-
C8J_1298, a bifunctional thiol oxidoreductase of Campylobacter jejuni, affects Dsb (disulfide bond) network functioning.PLoS One. 2020 Mar 23;15(3):e0230366. doi: 10.1371/journal.pone.0230366. eCollection 2020. PLoS One. 2020. PMID: 32203539 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

