Phenome-Wide Association Study to Explore Relationships between Immune System Related Genetic Loci and Complex Traits and Diseases
- PMID: 27508393
- PMCID: PMC4980020
- DOI: 10.1371/journal.pone.0160573
Phenome-Wide Association Study to Explore Relationships between Immune System Related Genetic Loci and Complex Traits and Diseases
Abstract
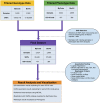
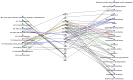
We performed a Phenome-Wide Association Study (PheWAS) to identify interrelationships between the immune system genetic architecture and a wide array of phenotypes from two de-identified electronic health record (EHR) biorepositories. We selected variants within genes encoding critical factors in the immune system and variants with known associations with autoimmunity. To define case/control status for EHR diagnoses, we used International Classification of Diseases, Ninth Revision (ICD-9) diagnosis codes from 3,024 Geisinger Clinic MyCode® subjects (470 diagnoses) and 2,899 Vanderbilt University Medical Center BioVU biorepository subjects (380 diagnoses). A pooled-analysis was also carried out for the replicating results of the two data sets. We identified new associations with potential biological relevance including SNPs in tumor necrosis factor (TNF) and ankyrin-related genes associated with acute and chronic sinusitis and acute respiratory tract infection. The two most significant associations identified were for the C6orf10 SNP rs6910071 and "rheumatoid arthritis" (ICD-9 code category 714) (pMETAL = 2.58 x 10-9) and the ATN1 SNP rs2239167 and "diabetes mellitus, type 2" (ICD-9 code category 250) (pMETAL = 6.39 x 10-9). This study highlights the utility of using PheWAS in conjunction with EHRs to discover new genotypic-phenotypic associations for immune-system related genetic loci.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

