Intrinsic protein disorder could be overlooked in cocrystallization conditions: An SRCD case study
- PMID: 27508941
- PMCID: PMC5079248
- DOI: 10.1002/pro.3010
Intrinsic protein disorder could be overlooked in cocrystallization conditions: An SRCD case study
Abstract
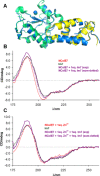
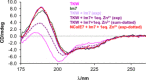
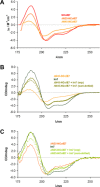
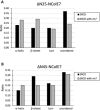
X-ray diffractometry dominates protein studies, as it can provide 3D structures of these diverse macromolecules or their molecular complexes with interacting partners: substrates, inhibitors, and/or cofactors. Here, we show that under cocrystallization conditions the results could reflect induced protein folds instead of the (partially) disordered original structures. The analysis of synchrotron radiation circular dichroism spectra revealed that the Im7 immunity protein stabilizes the native-like solution structure of unfolded NColE7 nuclease mutants via complex formation. This is consistent with the fact that among the several available crystal structures with its inhibitor or substrate, all NColE7 structures are virtually the same. Our results draw attention to the possible structural consequence of protein modifications, which is often hidden by compensational effects of intermolecular interactions. The growing evidence on the importance of protein intrinsic disorder thus, demands more extensive complementary experiments in solution phase with the unligated form of the protein of interest.
Keywords: Im7 immunity protein; SRCD spectroscopy; colicin E7 nuclease; induced protein folding.
© 2016 The Protein Society.
Figures



References
-
- Németh E, Körtvélyesi T, Kožíšek M, Thulstrup PW, Christensen HEM, Asaka MN, Nagata K, Gyurcsik B (2014) Substrate binding activates the designed triple mutant of the colicin E7 metallonuclease. J Biol Inorg Chem 19:1295–1303. - PubMed
-
- Németh E, Kožíšek M, Schilli GK, Gyurcsik B (2015) Preorganization of the catalytic Zn2+‐binding site in the HNH nuclease motif—a solution study. J Inorg Biochem 151:143–149. - PubMed
-
- Papadakos G, Sharma A, Lancaster LE, Bowen R, Kaminska R, Leech AP, Walker D, Redfield C, Kleanthous C (2015) Consequences of inducing intrinsic disorder in a high‐affinity protein−protein interaction. J Am Chem Soc 137:5252–5255. - PubMed
-
- Chak KF, Kuo WS, Lu FM, James R (1991) Cloning and characterization of the ColE7 plasmid. J Gen Microbiol 137:91–100. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

