Molecular basis of APC/C regulation by the spindle assembly checkpoint
- PMID: 27509861
- PMCID: PMC5019344
- DOI: 10.1038/nature19083
Molecular basis of APC/C regulation by the spindle assembly checkpoint
Abstract
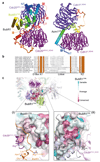
In the dividing eukaryotic cell, the spindle assembly checkpoint (SAC) ensures that each daughter cell inherits an identical set of chromosomes. The SAC coordinates the correct attachment of sister chromatid kinetochores to the mitotic spindle with activation of the anaphase-promoting complex (APC/C), the E3 ubiquitin ligase responsible for initiating chromosome separation. In response to unattached kinetochores, the SAC generates the mitotic checkpoint complex (MCC), which inhibits the APC/C and delays chromosome segregation. By cryo-electron microscopy, here we determine the near-atomic resolution structure of a human APC/C–MCC complex (APC/C(MCC)). Degron-like sequences of the MCC subunit BubR1 block degron recognition sites on Cdc20, the APC/C coactivator subunit responsible for substrate interactions. BubR1 also obstructs binding of the initiating E2 enzyme UbcH10 to repress APC/C ubiquitination activity. Conformational variability of the complex enables UbcH10 association, and structural analysis shows how the Cdc20 subunit intrinsic to the MCC (Cdc20(MCC)) is ubiquitinated, a process that results in APC/C reactivation when the SAC is silenced.
Conflict of interest statement
The authors declare no competing financial interests.
Figures











Comment in
-
Cell division: Mitotic regulation comes into focus.Nature. 2016 Aug 25;536(7617):407-8. doi: 10.1038/nature19423. Epub 2016 Aug 10. Nature. 2016. PMID: 27509855 No abstract available.
References
-
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Current biology: CB. 2012;22:R966–980. - PubMed
-
- Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Current biology : CB. 2015;25:R1002–1018. - PubMed
-
- Hoyt MA, Totis L, Roberts BTS. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

