Membranome: a database for proteome-wide analysis of single-pass membrane proteins
- PMID: 27510400
- PMCID: PMC5210604
- DOI: 10.1093/nar/gkw712
Membranome: a database for proteome-wide analysis of single-pass membrane proteins
Abstract
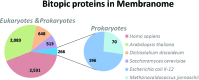
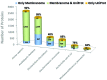
The Membranome database was developed to assist analysis and computational modeling of single-pass (bitopic) transmembrane (TM) proteins and their complexes by providing structural information about these proteins on a genomic scale. The database currently collects data on >6000 bitopic proteins from Homo sapiens, Arabidopsis thaliana, Dictyostelium discoideum, Saccharomyces cerevisiae, Escherichia coli and Methanocaldococcus jannaschii It presents the following data: (i) hierarchical classification of bitopic proteins into 15 functional classes, 689 structural superfamilies and 1404 families; (ii) 446 complexes of bitopic proteins with known three-dimensional (3D) structures classified into 129 families; (iii) computationally generated three-dimensional models of TM α-helices positioned in membranes; (iv) amino acid sequences, domain architecture, functional annotation and available experimental structures of bitopic proteins; (v) TM topology and intracellular localization, (vi) physical interactions between proteins from the database along with links to other resources. The database is freely accessible at http://membranome.org There is a variety of options for browsing, sorting, searching and retrieval of the content, including downloadable coordinate files of TM domains with calculated membrane boundaries.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Membranome 2.0: database for proteome-wide profiling of bitopic proteins and their dimers.Bioinformatics. 2018 Mar 15;34(6):1061-1062. doi: 10.1093/bioinformatics/btx720. Bioinformatics. 2018. PMID: 29126305
-
Membranome 3.0: Database of single-pass membrane proteins with AlphaFold models.Protein Sci. 2022 May;31(5):e4318. doi: 10.1002/pro.4318. Protein Sci. 2022. PMID: 35481632 Free PMC article.
-
Evolution and adaptation of single-pass transmembrane proteins.Biochim Biophys Acta Biomembr. 2018 Feb;1860(2):364-377. doi: 10.1016/j.bbamem.2017.11.002. Epub 2017 Nov 10. Biochim Biophys Acta Biomembr. 2018. PMID: 29129605
-
Database Search Engines: Paradigms, Challenges and Solutions.Adv Exp Med Biol. 2016;919:147-156. doi: 10.1007/978-3-319-41448-5_6. Adv Exp Med Biol. 2016. PMID: 27975215 Review.
-
Useful Web Resources.Adv Exp Med Biol. 2016;919:249-253. doi: 10.1007/978-3-319-41448-5_14. Adv Exp Med Biol. 2016. PMID: 27975223 Review.
Cited by
-
Polypharmacological Cell-Penetrating Peptides from Venomous Marine Animals Based on Immunomodulating, Antimicrobial, and Anticancer Properties.Mar Drugs. 2022 Dec 4;20(12):763. doi: 10.3390/md20120763. Mar Drugs. 2022. PMID: 36547910 Free PMC article.
-
Multiscale Simulations of Biological Membranes: The Challenge To Understand Biological Phenomena in a Living Substance.Chem Rev. 2019 May 8;119(9):5607-5774. doi: 10.1021/acs.chemrev.8b00538. Epub 2019 Mar 12. Chem Rev. 2019. PMID: 30859819 Free PMC article.
-
Spiers Memorial Lecture: Analysis and de novo design of membrane-interactive peptides.Faraday Discuss. 2021 Dec 24;232(0):9-48. doi: 10.1039/d1fd00061f. Faraday Discuss. 2021. PMID: 34693965 Free PMC article.
-
Transmembrane Membrane Readers form a Novel Class of Proteins That Include Peripheral Phosphoinositide Recognition Domains and Viral Spikes.Membranes (Basel). 2022 Nov 17;12(11):1161. doi: 10.3390/membranes12111161. Membranes (Basel). 2022. PMID: 36422153 Free PMC article. Review.
-
Comparison of Current Methods for Signal Peptide Prediction in Phytoplasmas.Front Microbiol. 2021 Mar 25;12:661524. doi: 10.3389/fmicb.2021.661524. eCollection 2021. Front Microbiol. 2021. PMID: 33841387 Free PMC article.
References
-
- Reeb J., Kloppmann E., Bernhofer M., Rost B. Evaluation of transmembrane helix predictions in 2014. Proteins. 2015;83:473–484. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

