Assuring the Quality of Next-Generation Sequencing in Clinical Microbiology and Public Health Laboratories
- PMID: 27510831
- PMCID: PMC5121372
- DOI: 10.1128/JCM.00949-16
Assuring the Quality of Next-Generation Sequencing in Clinical Microbiology and Public Health Laboratories
Abstract
Clinical microbiology and public health laboratories are beginning to utilize next-generation sequencing (NGS) for a range of applications. This technology has the potential to transform the field by providing approaches that will complement, or even replace, many conventional laboratory tests. While the benefits of NGS are significant, the complexities of these assays require an evolving set of standards to ensure testing quality. Regulatory and accreditation requirements, professional guidelines, and best practices that help ensure the quality of NGS-based tests are emerging. This review highlights currently available standards and guidelines for the implementation of NGS in the clinical and public health laboratory setting, and it includes considerations for NGS test validation, quality control procedures, proficiency testing, and reference materials.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
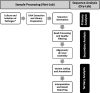


References
-
- Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, Bouquet J, Greninger AL, Luk KC, Enge B, Wadford DA, Messenger SL, Genrich GL, Pellegrino K, Grard G, Leroy E, Schneider BS, Fair JN, Martinez MA, Isa P, Crump JA, DeRisi JL, Sittler T, Hackett J Jr, Miller S, Chiu CY. 2014. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 24:1180–1192. doi:10.1101/gr.171934.113. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

