Quorum sensing signal-response systems in Gram-negative bacteria
- PMID: 27510864
- PMCID: PMC5056591
- DOI: 10.1038/nrmicro.2016.89
Quorum sensing signal-response systems in Gram-negative bacteria
Abstract
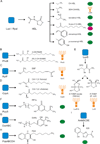
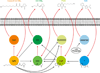
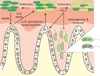
Bacteria use quorum sensing to orchestrate gene expression programmes that underlie collective behaviours. Quorum sensing relies on the production, release, detection and group-level response to extracellular signalling molecules, which are called autoinducers. Recent work has discovered new autoinducers in Gram-negative bacteria, shown how these molecules are recognized by cognate receptors, revealed new regulatory components that are embedded in canonical signalling circuits and identified novel regulatory network designs. In this Review we examine how, together, these features of quorum sensing signal-response systems combine to control collective behaviours in Gram-negative bacteria and we discuss the implications for host-microbial associations and antibacterial therapy.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

