Ribonucleotides and Transcription-Associated Mutagenesis in Yeast
- PMID: 27511624
- PMCID: PMC5296399
- DOI: 10.1016/j.jmb.2016.08.005
Ribonucleotides and Transcription-Associated Mutagenesis in Yeast
Abstract
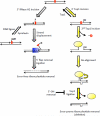
High levels of transcription stimulate mutation rates in microorganisms, and this occurs primarily through an enhanced accumulation of DNA damage. The major source of transcription-associated damage in yeast is Topoisomerase I (Top1), an enzyme that removes torsional stress that accumulates when DNA strands are separated. Top1 relieves torsional stress by nicking and resealing one DNA strand, and some Top1-dependent mutations are due to trapping and processing of the covalent cleavage intermediate. Most, however, reflect enzyme incision at ribonucleotides, which are the most abundant noncanonical component of DNA. In either case, Top1 generates a distinctive mutation signature composed of short deletions in tandem repeats; in the specific case of ribonucleotide-initiated events, mutations reflect sequential cleavage by the enzyme. Top1-dependent mutations do not require highly activated transcription, but their levels are greatly increased by transcription, which partially reflects an interaction of Top1 with RNA polymerase. Recent studies have demonstrated that Top1-dependent mutations exhibit a strand bias, with the nature of the bias differing depending on the transcriptional status of the underlying DNA. Under low-transcription conditions, most Top1-dependent mutations arise in the context of replication and reflect incision at ribonucleotides incorporated during leading-strand synthesis. Under high-transcription conditions, most Top1-dependent events arise when the enzyme cleaves the non-transcribed strand of DNA. In addition to increasing genetic instability in growing cells, Top1 activity in transcriptionally active regions may be a source of mutations in quiescent cells.
Keywords: Topoisomerase 1; mutagenesis; ribonucleotides; transcription.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
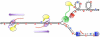


Similar articles
-
Studying Topoisomerase 1-Mediated Damage at Genomic Ribonucleotides.Methods Mol Biol. 2018;1703:241-257. doi: 10.1007/978-1-4939-7459-7_17. Methods Mol Biol. 2018. PMID: 29177746
-
Parallel analysis of ribonucleotide-dependent deletions produced by yeast Top1 in vitro and in vivo.Nucleic Acids Res. 2016 Sep 19;44(16):7714-21. doi: 10.1093/nar/gkw495. Epub 2016 Jun 1. Nucleic Acids Res. 2016. PMID: 27257064 Free PMC article.
-
Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast.DNA Repair (Amst). 2013 Mar 1;12(3):205-11. doi: 10.1016/j.dnarep.2012.12.004. Epub 2013 Jan 8. DNA Repair (Amst). 2013. PMID: 23305949 Free PMC article.
-
The Top1 paradox: Friend and foe of the eukaryotic genome.DNA Repair (Amst). 2017 Aug;56:33-41. doi: 10.1016/j.dnarep.2017.06.005. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28641942 Free PMC article. Review.
-
Genome instabilities arising from ribonucleotides in DNA.DNA Repair (Amst). 2017 Aug;56:26-32. doi: 10.1016/j.dnarep.2017.06.004. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28629774 Free PMC article. Review.
Cited by
-
RNases H1 and H2: guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis.Curr Genet. 2020 Dec;66(6):1073-1084. doi: 10.1007/s00294-020-01086-8. Epub 2020 Sep 4. Curr Genet. 2020. PMID: 32886170 Free PMC article. Review.
-
Topoisomerase 1 facilitates nucleosome reassembly at stress genes during recovery.Nucleic Acids Res. 2023 Dec 11;51(22):12161-12173. doi: 10.1093/nar/gkad1066. Nucleic Acids Res. 2023. PMID: 37956308 Free PMC article.
-
The divergence of mutation rates and spectra across the Tree of Life.EMBO Rep. 2023 Oct 9;24(10):e57561. doi: 10.15252/embr.202357561. Epub 2023 Aug 24. EMBO Rep. 2023. PMID: 37615267 Free PMC article. Review.
-
Both R-loop removal and ribonucleotide excision repair activities of RNase H2 contribute substantially to chromosome stability.DNA Repair (Amst). 2017 Apr;52:110-114. doi: 10.1016/j.dnarep.2017.02.012. Epub 2017 Feb 20. DNA Repair (Amst). 2017. PMID: 28268090 Free PMC article.
-
Ribonucleotide Incorporation by Eukaryotic B-Family Replicases and Its Implications for Genome Stability.Annu Rev Biochem. 2022 Jun 21;91:133-155. doi: 10.1146/annurev-biochem-032620-110354. Epub 2022 Mar 14. Annu Rev Biochem. 2022. PMID: 35287470 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

