Systems Analysis of the Complement-Induced Priming Phase of Liver Regeneration
- PMID: 27511733
- PMCID: PMC5038987
- DOI: 10.4049/jimmunol.1600628
Systems Analysis of the Complement-Induced Priming Phase of Liver Regeneration
Abstract
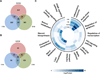
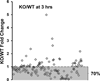
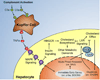
Liver regeneration is a well-orchestrated process in the liver that allows mature hepatocytes to reenter the cell cycle to proliferate and replace lost or damaged cells. This process is often impaired in fatty or diseased livers, leading to cirrhosis and other deleterious phenotypes. Prior research has established the role of the complement system and its effector proteins in the progression of liver regeneration; however, a detailed mechanistic understanding of the involvement of complement in regeneration is yet to be established. In this study, we have examined the role of the complement system during the priming phase of liver regeneration through a systems level analysis using a combination of transcriptomic and metabolomic measurements. More specifically, we have performed partial hepatectomy on mice with genetic deficiency in C3, the major component of the complement cascade, and collected their livers at various time points. Based on our analysis, we show that the C3 cascade activates c-fos and promotes the TNF-α signaling pathway, which then activates acute-phase genes such as serum amyloid proteins and orosomucoids. The complement activation also regulates the efflux and the metabolism of cholesterol, an important metabolite for cell cycle and proliferation. Based on our systems level analysis, we provide an integrated model for the complement-induced priming phase of liver regeneration.
Copyright © 2016 by The American Association of Immunologists, Inc.
Figures



References
-
- Clavien PA. Liver regeneration: a spotlight on the novel role of platelets and serotonin. Swiss medical weekly. 2008;138:361–370. - PubMed
-
- Clavien PA, Petrowsky H, DeOliveira ML, Graf R. Strategies for safer liver surgery and partial liver transplantation. The New England journal of medicine. 2007;356:1545–1559. - PubMed
-
- Gotohda N, Iwagaki H, Ozaki M, Kinoshita T, Konishi M, Nakagohri T, Takahashi S, Saito S, Yagi T, Tanaka N. Deficient response of IL-6 impaired liver regeneration after hepatectomy in patients with viral hepatitis. Hepato-gastroenterology. 2008;55:1439–1444. - PubMed
-
- Hines IN, Kremer M, Isayama F, Perry AW, Milton RJ, Black AL, Byrd CL, Wheeler MD. Impaired liver regeneration and increased oval cell numbers following T cell-mediated hepatitis. Hepatology. 2007;46:229–241. - PubMed
-
- Tanemura A, Mizuno S, Wada H, Yamada T, Nobori T, Isaji S. Donor age affects liver regeneration during early period in the graft liver and late period in the remnant liver after living donor liver transplantation. World journal of surgery. 2012;36:1102–1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

