FAM134B, the Selective Autophagy Receptor for Endoplasmic Reticulum Turnover, Inhibits Replication of Ebola Virus Strains Makona and Mayinga
- PMID: 27511895
- PMCID: PMC5050481
- DOI: 10.1093/infdis/jiw270
FAM134B, the Selective Autophagy Receptor for Endoplasmic Reticulum Turnover, Inhibits Replication of Ebola Virus Strains Makona and Mayinga
Abstract
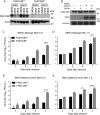
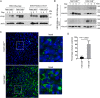
Selective autophagy of the endoplasmic reticulum (termed ER-phagy) is controlled by members of the FAM134 reticulon protein family. Here we used mouse embryonic fibroblasts from mice deficient in FAM134B to examine the role of the ER in replication of historic (Mayinga) or contemporary (Makona GCO7) strains of Ebola virus (EBOV). Loss of FAM134B resulted in 1-2 log10 higher production of infectious EBOV, which was associated with increased production of viral proteins GP and VP40 and greater accumulation of nucleocaspid lattices. In addition, only 10% of wild-type cells contained detectable nucleoprotein, whereas knockout of FAM134B resulted in 80% of cells positive for nucleoprotein. Together, these data suggest that FAM134B-dependent ER-phagy is an important limiting event in EBOV replication in mouse cells and may have implications for further development of antiviral therapeutics and murine models of infection.
Keywords: ER-phagy; Ebolavirus; FAM134B; mouse models; reticulon; selective autophagy; virus replication.
Published by Oxford University Press for the Infectious Diseases Society of America 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures
References
-
- Bah EI, Lamah MC, Fletcher T et al. . Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. N Engl J Med 2015; 372:40–7. - PubMed
-
- Chan SY, Ma MC, Goldsmith MA. Differential induction of cellular detachment by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J Gen Virol 2000; 81:2155–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

