Benign Rabbit Caliciviruses Exhibit Evolutionary Dynamics Similar to Those of Their Virulent Relatives
- PMID: 27512059
- PMCID: PMC5044836
- DOI: 10.1128/JVI.01212-16
Benign Rabbit Caliciviruses Exhibit Evolutionary Dynamics Similar to Those of Their Virulent Relatives
Abstract
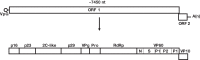
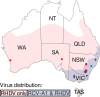
Two closely related caliciviruses cocirculate in Australia: rabbit hemorrhagic disease virus (RHDV) and rabbit calicivirus Australia 1 (RCV-A1). RCV-A1 causes benign enteric infections in the European rabbit (Oryctolagus cuniculus) in Australia and New Zealand, while its close relative RHDV causes a highly pathogenic infection of the liver in the same host. The comparison of these viruses provides important information on the nature and trajectory of virulence evolution, particularly as highly virulent strains of RHDV may have evolved from nonpathogenic ancestors such as RCV-A1. To determine the evolution of RCV-A1 we sequenced the full-length genomes of 44 RCV-A1 samples isolated from healthy rabbits and compared key evolutionary parameters to those of its virulent relative, RHDV. Despite their marked differences in pathogenicity and tissue tropism, RCV-A1 and RHDV have evolved in a very similar manner. Both viruses have evolved at broadly similar rates, suggesting that their dynamics are largely shaped by high background mutation rates, and both exhibit occasional recombination and an evolutionary environment dominated by purifying selection. In addition, our comparative analysis revealed that there have been multiple changes in both virulence and tissue tropism in the evolutionary history of these and related viruses. Finally, these new genomic data suggest that either RCV-A1 was introduced into Australia after the introduction of myxoma virus as a biocontrol agent in 1950 or there was drastic reduction of the rabbit population, and hence of RCV-A1 genetic diversity, perhaps coincident with the emergence of myxoma virus.
Importance: The comparison of closely related viruses that differ profoundly in propensity to cause disease in their hosts offers a powerful opportunity to reveal the causes of changes in virulence and to study how such changes alter the evolutionary dynamics of these pathogens. Here we describe such a novel comparison involving two closely related RNA viruses that cocirculate in Australia, the highly virulent rabbit hemorrhagic disease virus (RHDV) and the nonpathogenic rabbit calicivirus Australia 1 (RCV-A1). Both viruses infect the European rabbit, but they differ in virulence, tissue tropism, and mechanisms of transmission. Surprisingly, and despite these fundamental differences, RCV-A1 and RHDV have evolved at very similar (high) rates and with strong purifying selection. Furthermore, candidate key mutations were identified that may play a role in virulence and/or tissue tropism and therefore warrant further investigation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Characterisation of Lagovirus europaeus GI-RHDVs (Rabbit Haemorrhagic Disease Viruses) in Terms of Their Pathogenicity and Immunogenicity.Int J Mol Sci. 2024 May 14;25(10):5342. doi: 10.3390/ijms25105342. Int J Mol Sci. 2024. PMID: 38791380 Free PMC article. Review.
-
Benign Rabbit Calicivirus in New Zealand.Appl Environ Microbiol. 2017 May 17;83(11):e00090-17. doi: 10.1128/AEM.00090-17. Print 2017 Jun 1. Appl Environ Microbiol. 2017. PMID: 28363968 Free PMC article.
-
DIFFERENT SEROLOGICAL PROFILES TO CO-OCCURRING PATHOGENIC AND NONPATHOGENIC CALICIVIRUSES IN WILD EUROPEAN RABBITS (ORYCTOLAGUS CUNICULUS) ACROSS AUSTRALIA.J Wildl Dis. 2017 Jul;53(3):472-481. doi: 10.7589/2016-06-148. Epub 2017 Feb 23. J Wildl Dis. 2017. PMID: 28231031
-
Distribution and prevalence of the Australian non-pathogenic rabbit calicivirus is correlated with rainfall and temperature.PLoS One. 2014 Dec 8;9(12):e113976. doi: 10.1371/journal.pone.0113976. eCollection 2014. PLoS One. 2014. PMID: 25486092 Free PMC article.
-
Viral biocontrol: grand experiments in disease emergence and evolution.Trends Microbiol. 2015 Feb;23(2):83-90. doi: 10.1016/j.tim.2014.10.004. Epub 2014 Oct 31. Trends Microbiol. 2015. PMID: 25455418 Free PMC article. Review.
Cited by
-
The discovery of three new hare lagoviruses reveals unexplored viral diversity in this genus.Virus Evol. 2019 Apr 9;5(1):vez005. doi: 10.1093/ve/vez005. eCollection 2019 Jan. Virus Evol. 2019. PMID: 30997155 Free PMC article.
-
Characterization of old RHDV strains by complete genome sequencing identifies a novel genetic group.Sci Rep. 2017 Oct 19;7(1):13599. doi: 10.1038/s41598-017-13902-2. Sci Rep. 2017. PMID: 29051566 Free PMC article.
-
Characterisation of Lagovirus europaeus GI-RHDVs (Rabbit Haemorrhagic Disease Viruses) in Terms of Their Pathogenicity and Immunogenicity.Int J Mol Sci. 2024 May 14;25(10):5342. doi: 10.3390/ijms25105342. Int J Mol Sci. 2024. PMID: 38791380 Free PMC article. Review.
-
Genetic Characteristics and Phylogeographic Dynamics of Lagoviruses, 1988-2021.Viruses. 2023 Mar 23;15(4):815. doi: 10.3390/v15040815. Viruses. 2023. PMID: 37112796 Free PMC article.
-
Retrospective Analysis Shows That Most RHDV GI.1 Strains Circulating Since the Late 1990s in France and Sweden Were Recombinant GI.3P-GI.1d Strains.Genes (Basel). 2020 Aug 9;11(8):910. doi: 10.3390/genes11080910. Genes (Basel). 2020. PMID: 32784857 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

