Novel Acylguanidine-Based Inhibitor of HIV-1
- PMID: 27512074
- PMCID: PMC5044834
- DOI: 10.1128/JVI.01107-16
Novel Acylguanidine-Based Inhibitor of HIV-1
Abstract
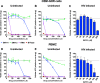
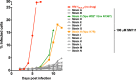
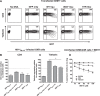
The emergence of transmissible HIV-1 strains with resistance to antiretroviral drugs highlights a continual need for new therapies. Here we describe a novel acylguanidine-containing compound, 1-(2-(azepan-1-yl)nicotinoyl)guanidine (or SM111), that inhibits in vitro replication of HIV-1, including strains resistant to licensed protease, reverse transcriptase, and integrase inhibitors, without major cellular toxicity. At inhibitory concentrations, intracellular p24(Gag) production was unaffected, but virion release (measured as extracellular p24(Gag)) was reduced and virion infectivity was substantially impaired, suggesting that SM111 acts at a late stage of viral replication. SM111-mediated inhibition of HIV-1 was partially overcome by a Vpu I17R mutation alone or a Vpu W22* truncation in combination with Env N136Y. These mutations enhanced virion infectivity and Env expression on the surface of infected cells in the absence and presence of SM111 but also impaired Vpu's ability to downregulate CD4 and BST2/tetherin. Taken together, our results support acylguanidines as a class of HIV-1 inhibitors with a distinct mechanism of action compared to that of licensed antiretrovirals. Further research on SM111 and similar compounds may help to elucidate knowledge gaps related to Vpu's role in promoting viral egress and infectivity.
Importance: New inhibitors of HIV-1 replication may be useful as therapeutics to counteract drug resistance and as reagents to perform more detailed studies of viral pathogenesis. SM111 is a small molecule that blocks the replication of wild-type and drug-resistant HIV-1 strains by impairing viral release and substantially reducing virion infectivity, most likely through its ability to prevent Env expression at the infected cell surface. Partial resistance to SM111 is mediated by mutations in Vpu and/or Env, suggesting that the compound affects host/viral protein interactions that are important during viral egress. Further characterization of SM111 and similar compounds may allow more detailed pharmacological studies of HIV-1 egress and provide opportunities to develop new treatments for HIV-1.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
Preadaptation of Simian Immunodeficiency Virus SIVsmm Facilitated Env-Mediated Counteraction of Human Tetherin by Human Immunodeficiency Virus Type 2.J Virol. 2018 Aug 29;92(18):e00276-18. doi: 10.1128/JVI.00276-18. Print 2018 Sep 15. J Virol. 2018. PMID: 29976668 Free PMC article.
-
Activities of transmitted/founder and chronic clade B HIV-1 Vpu and a C-terminal polymorphism specifically affecting virion release.J Virol. 2014 May;88(9):5062-78. doi: 10.1128/JVI.03472-13. Epub 2014 Feb 26. J Virol. 2014. PMID: 24574397 Free PMC article.
-
Glycosyl-Phosphatidylinositol-Anchored Anti-HIV Env Single-Chain Variable Fragments Interfere with HIV-1 Env Processing and Viral Infectivity.J Virol. 2018 Mar 14;92(7):e02080-17. doi: 10.1128/JVI.02080-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29321330 Free PMC article.
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
-
Targets for inhibition of HIV replication: entry, enzyme action, release and maturation.Intervirology. 2012;55(2):84-97. doi: 10.1159/000331995. Epub 2012 Jan 24. Intervirology. 2012. PMID: 22286875 Review.
Cited by
-
Various plus unique: Viral protein U as a plurifunctional protein for HIV-1 replication.Exp Biol Med (Maywood). 2017 Apr;242(8):850-858. doi: 10.1177/1535370217697384. Epub 2017 Jan 1. Exp Biol Med (Maywood). 2017. PMID: 28346011 Free PMC article. Review.
-
Ammonium Formate-Pd/C as a New Reducing System for 1,2,4-Oxadiazoles. Synthesis of Guanidine Derivatives and Reductive Rearrangement to Quinazolin-4-Ones with Potential Anti-Diabetic Activity.Int J Mol Sci. 2021 Nov 14;22(22):12301. doi: 10.3390/ijms222212301. Int J Mol Sci. 2021. PMID: 34830187 Free PMC article.
-
2-Trifluoromethylthiazole-5-carboxamides: Analogues of a Stilbene-Based Anti-HIV Agent that Impact HIV mRNA Processing.ACS Med Chem Lett. 2021 Oct 29;12(11):1818-1823. doi: 10.1021/acsmedchemlett.1c00428. eCollection 2021 Nov 11. ACS Med Chem Lett. 2021. PMID: 34795872 Free PMC article.
-
Flavonoid-based inhibition of cyclin-dependent kinase 9 without concomitant inhibition of histone deacetylases durably reinforces HIV latency.Biochem Pharmacol. 2021 Apr;186:114462. doi: 10.1016/j.bcp.2021.114462. Epub 2021 Feb 10. Biochem Pharmacol. 2021. PMID: 33577894 Free PMC article.
-
High-Throughput NanoBiT-Based Screening for Inhibitors of HIV-1 Vpu and Host BST-2 Protein Interaction.Int J Mol Sci. 2021 Aug 27;22(17):9308. doi: 10.3390/ijms22179308. Int J Mol Sci. 2021. PMID: 34502213 Free PMC article.
References
-
- UNAIDS. 2013. UNAIDS report on the global AIDS epidemic. UNAIDS, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

