Circadian Oscillation of the Lettuce Transcriptome under Constant Light and Light-Dark Conditions
- PMID: 27512400
- PMCID: PMC4961695
- DOI: 10.3389/fpls.2016.01114
Circadian Oscillation of the Lettuce Transcriptome under Constant Light and Light-Dark Conditions
Abstract
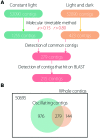
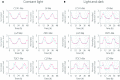
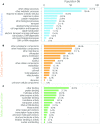
Although, the circadian clock is a universal biological system in plants and it orchestrates important role of plant production such as photosynthesis, floral induction and growth, there are few such studies on cultivated species. Lettuce is one major cultivated species for both open culture and plant factories and there is little information concerning its circadian clock system. In addition, most of the relevant genes have not been identified. In this study, we detected circadian oscillation in the lettuce transcriptome using time-course RNA sequencing (RNA-Seq) data. Constant light (LL) and light-dark (LD) conditions were used to detect circadian oscillation because the circadian clock has some basic properties: one is self-sustaining oscillation under constant light and another is entrainment to environmental cycles such as light and temperature. In the results, 215 contigs were detected as common oscillating contigs under both LL and LD conditions. The 215 common oscillating contigs included clock gene-like contigs CCA1 (CIRCADIAN CLOCK ASSOCIATED 1)-like, TOC1 (TIMING OF CAB EXPRESSION 1)-like and LHY (LATE ELONGATED HYPOCOTYL)-like, and their expression patterns were similar to those of Arabidopsis. Functional enrichment analysis by GO (gene ontology) Slim and GO Fat showed that the GO terms of response to light stimulus, response to stress, photosynthesis and circadian rhythms were enriched in the 215 common oscillating contigs and these terms were actually regulated by circadian clocks in plants. The 215 common oscillating contigs can be used to evaluate whether the gene expression pattern related to photosynthesis and optical response performs normally in lettuce.
Keywords: circadian clocks; enrichment; lettuce; oscillations; plant factories; transcriptome.
Figures


Similar articles
-
LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis.BMC Plant Biol. 2016 May 20;16(1):114. doi: 10.1186/s12870-016-0810-8. BMC Plant Biol. 2016. PMID: 27207270 Free PMC article.
-
ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY.Plant J. 2005 Oct;44(2):300-13. doi: 10.1111/j.1365-313X.2005.02531.x. Plant J. 2005. PMID: 16212608
-
Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression.FEBS Lett. 2004 Jan 16;557(1-3):259-64. doi: 10.1016/s0014-5793(03)01470-4. FEBS Lett. 2004. PMID: 14741378
-
MYB transcription factors in the Arabidopsis circadian clock.J Exp Bot. 2002 Jul;53(374):1551-7. doi: 10.1093/jxb/erf027. J Exp Bot. 2002. PMID: 12096093 Review.
-
The circadian system of Arabidopsis thaliana: forward and reverse genetic approaches.Chronobiol Int. 1999 Jan;16(1):1-16. doi: 10.3109/07420529908998708. Chronobiol Int. 1999. PMID: 10023572 Review.
Cited by
-
Identification of the global diurnal rhythmic transcripts, transcription factors and time-of-day specific cis elements in Chenopodium quinoa.BMC Plant Biol. 2023 Feb 16;23(1):96. doi: 10.1186/s12870-023-04107-z. BMC Plant Biol. 2023. PMID: 36793005 Free PMC article.
-
Transcriptome Analysis of Diurnal Gene Expression in Chinese Cabbage.Genes (Basel). 2019 Feb 11;10(2):130. doi: 10.3390/genes10020130. Genes (Basel). 2019. PMID: 30754711 Free PMC article.
-
Translating Flowering Time From Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species.Plants (Basel). 2018 Dec 16;7(4):111. doi: 10.3390/plants7040111. Plants (Basel). 2018. PMID: 30558374 Free PMC article. Review.
-
A Composite Analysis of Flowering Time Regulation in Lettuce.Front Plant Sci. 2021 Mar 8;12:632708. doi: 10.3389/fpls.2021.632708. eCollection 2021. Front Plant Sci. 2021. PMID: 33763095 Free PMC article.
-
Metabolic Reprogramming in Leaf Lettuce Grown Under Different Light Quality and Intensity Conditions Using Narrow-Band LEDs.Sci Rep. 2018 May 21;8(1):7914. doi: 10.1038/s41598-018-25686-0. Sci Rep. 2018. PMID: 29784957 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

