The micromorphology of Trichoderma reesei analyzed in cultivations on lactose and solid lignocellulosic substrate, and its relationship with cellulase production
- PMID: 27512503
- PMCID: PMC4979124
- DOI: 10.1186/s13068-016-0584-0
The micromorphology of Trichoderma reesei analyzed in cultivations on lactose and solid lignocellulosic substrate, and its relationship with cellulase production
Abstract
Background: Trichoderma reesei is the principal producer of cellulolytic enzymes. Because of the strong influence on the enzyme production, the morphology of the filamentous fungi is a key parameter for process optimization. For cost-effective production of cellulolytic enzymes, the cultivation of T. reesei is performed on lignocellulosic waste streams. These insoluble substrates prevent the application of the conventional light microscopy for the analysis of fungal morphology. Here, we present a novel method for the micromorphological analysis based on confocal laser-scanning microscopy (CLSM) and the computer-aided image analysis. This method enabled the quantification of the dimensions of the single cell (intercalary length and cell width) and the degree of branching in cultivations on the industrially relevant substrates wheat straw and lactose. The micromorphology of two T. reesei strains, QM9414 and a carbon catabolite derepressed cre1 knockout mutant (Δcre1), was analyzed in dependence of substrate, inoculation method, and agitation velocity.
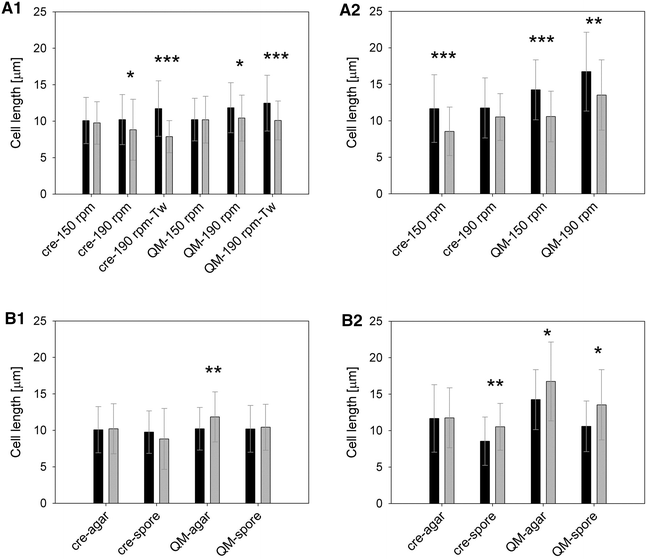
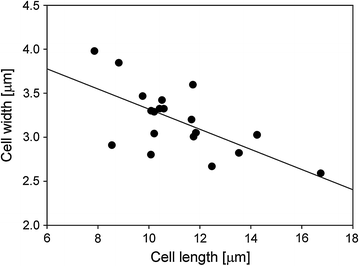
Results: Trichoderma reesei strain Δcre1 formed shorter cells (10.09 µm) on average and developed more ramified mycelia (0.36 branches/cell) than strain QM9414 (12.03 µm, 0.22 branches/cell). Cultivated on wheat straw, the average cell length of QM9414 (10.87 µm) and Δcre1 (9.74 µm) was 10 and 21 % shorter as compared to reference cultivations on lactose. When inoculation was done with spores as compared to hyphal biomass, cell lengths of QM9414 (10.97 µm) and Δcre1 (9.10 µm) were on average about 20 % shorter. Strain performance was evaluated in protein concentration and total cellulase activity, which varied between 0.69 and 2.31 FPU/mL for Δcre1 and between 0.84 and 1.64 FPU/mL for QM9414. The cell length exhibited slightly negative correlation with the protein (regression coefficient -0.04 g/(L µm), R (2) 0.33) and the cellulase (-0.30 FPU/(mL µm), R (2) 0.53) production.
Conclusions: The dimensions of the single cell of T. reesei were dependent on strain background, substrate used and process conditions applied. Micromorphological changes were correlated semi-quantitatively with the efficiency of enzyme production. In providing a process analytical tool for enzyme production by T. reesei on lignocellulosic substrate, this study has relevance for the characterization and optimization of a critical step in the overall saccharification process.
Keywords: Cellulase production; Lactose; Lignocellulose; Morphology; Trichoderma reesei; Wheat straw.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Optimizing microbioreactor cultivation strategies for Trichoderma reesei: from batch to fed-batch operations.Microb Cell Fact. 2024 Apr 15;23(1):112. doi: 10.1186/s12934-024-02371-8. Microb Cell Fact. 2024. PMID: 38622596 Free PMC article.
-
Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis.Fungal Genet Biol. 2014 Dec;73:93-103. doi: 10.1016/j.fgb.2014.10.009. Epub 2014 Oct 18. Fungal Genet Biol. 2014. PMID: 25459535
-
Production of the versatile cellulase for cellulose bioconversion and cellulase inducer synthesis by genetic improvement of Trichoderma reesei.Biotechnol Biofuels. 2017 Nov 15;10:272. doi: 10.1186/s13068-017-0963-1. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 29167702 Free PMC article.
-
Deciphering the molecular mechanisms behind cellulase production in Trichoderma reesei, the hyper-cellulolytic filamentous fungus.Biosci Biotechnol Biochem. 2016 Sep;80(9):1712-29. doi: 10.1080/09168451.2016.1171701. Epub 2016 Apr 14. Biosci Biotechnol Biochem. 2016. PMID: 27075508 Review.
-
An overview of Trichoderma reesei co-cultures for the production of lignocellulolytic enzymes.Appl Microbiol Biotechnol. 2021 Apr;105(8):3019-3025. doi: 10.1007/s00253-021-11261-7. Epub 2021 Apr 7. Appl Microbiol Biotechnol. 2021. PMID: 33825000 Review.
Cited by
-
Carbon catabolite repression involves physical interaction of the transcription factor CRE1/CreA and the Tup1-Cyc8 complex in Penicillium oxalicum and Trichoderma reesei.Biotechnol Biofuels. 2021 Dec 24;14(1):244. doi: 10.1186/s13068-021-02092-9. Biotechnol Biofuels. 2021. PMID: 34952627 Free PMC article.
-
Disruption of the Trichoderma reesei gul1 gene stimulates hyphal branching and reduces broth viscosity in cellulase production.J Ind Microbiol Biotechnol. 2021 Apr 30;48(1-2):kuab012. doi: 10.1093/jimb/kuab012. J Ind Microbiol Biotechnol. 2021. PMID: 33693788 Free PMC article.
-
Identification of chitin synthase activator in Aspergillus niger and its application in citric acid fermentation.Appl Microbiol Biotechnol. 2022 Nov;106(21):6993-7011. doi: 10.1007/s00253-022-12174-9. Epub 2022 Sep 23. Appl Microbiol Biotechnol. 2022. PMID: 36149454
-
The characteristics of insoluble softwood substrates affect fungal morphology, secretome composition, and hydrolytic efficiency of enzymes produced by Trichoderma reesei.Biotechnol Biofuels. 2021 Apr 26;14(1):105. doi: 10.1186/s13068-021-01955-5. Biotechnol Biofuels. 2021. PMID: 33902680 Free PMC article.
-
It Works! Organic-Waste-Assisted Trichoderma spp. Solid-State Fermentation on Agricultural Digestate.Microorganisms. 2022 Jan 13;10(1):164. doi: 10.3390/microorganisms10010164. Microorganisms. 2022. PMID: 35056614 Free PMC article.
References
-
- Sassner P, Galbe M, Zacchi G. Techno-economic evaluation of bioethanol production from three different lignocellulosic materials. Biomass Bioenergy. 2008;32(5):422–430. doi: 10.1016/j.biombioe.2007.10.014. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

